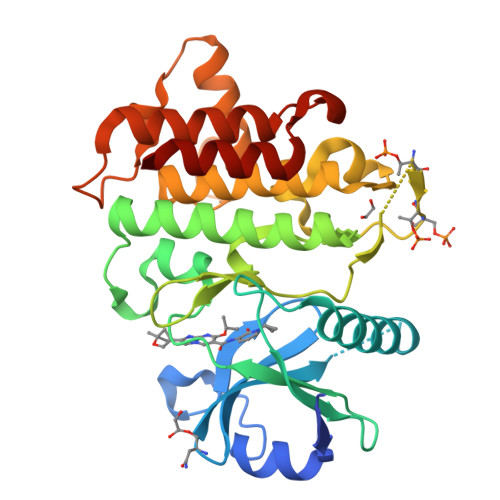
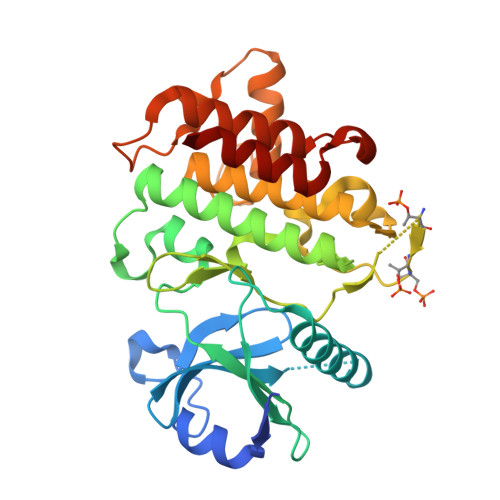
Discovery of BIO-8169─A Highly Potent, Selective, and Brain-Penetrant IRAK4 Inhibitor for the Treatment of Neuroinflammation.
Pfaffenbach, M., Bolduc, P.N., Xin, Z., Gao, F., Evans, R., Fang, T., Chodaparambil, J.V., Henry, K.L., Li, P., Mathieu, S., Metrick, C., Vera Rebollar, J.A., Gu, R.F., Mccarl, C.A., Silbereis, J., Peterson, E.A.(2024) J Med Chem 67: 8383-8395
- PubMed: 38695469
- DOI: https://doi.org/10.1021/acs.jmedchem.4c00560
- Primary Citation of Related Structures:
8TX0 - PubMed Abstract:
Interleukin receptor associated kinase 4 (IRAK4) plays an important role in innate immune signaling through Toll-like and interleukin-1 receptors and represents an attractive target for the treatment of inflammatory diseases and cancer. We previously reported the development of a potent, selective, and brain-penetrant imidazopyrimidine series of IRAK4 inhibitors. However, lead molecule BIO-7488 ( 1 ) suffered from low solubility which led to variable PK, compound accumulation, and poor in vivo tolerability. Herein, we describe the discovery of a series of pyridone analogs with improved solubility which are highly potent, selective and demonstrate desirable PK profiles including good oral bioavailability and excellent brain penetration. BIO-8169 ( 2 ) reduced the in vivo production of pro-inflammatory cytokines, was well tolerated in safety studies in rodents and dog at margins well above the predicted efficacious exposure and showed promising results in a mouse model for multiple sclerosis.
Organizational Affiliation:
Department of Medicinal Chemistry, Biogen Inc., 225 Binney Street, Cambridge, Massachusetts 02142, United States.