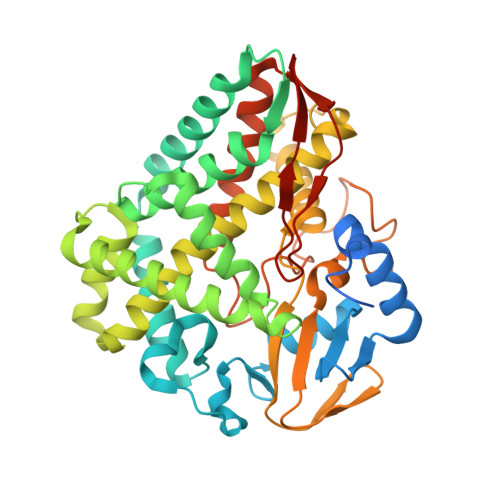
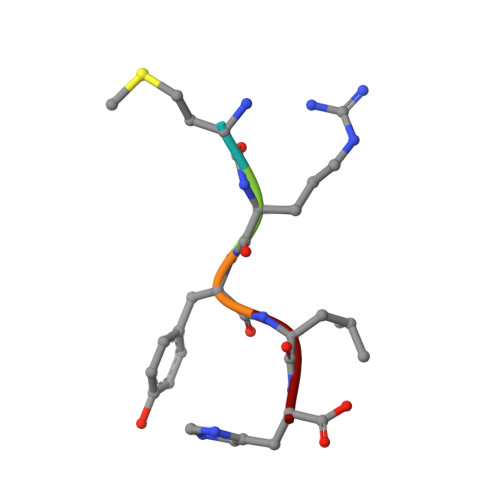
Structural Insights into a Side Chain Cross-Linking Biarylitide P450 from RiPP Biosynthesis
Hansen, M.H., Keto, A., Treisman, M., Sasi, V.M., Coe, L., Zhao, Y., Padva, L., Hess, C., Leichthammer, V., Machell, D.L., Schittenhelm, R.B., Jackson, C.J., Tailhades, J., Crusemann, M., De Voss, J.J., Krenske, E.H., Cryle, M.J.(2024) ACS Catal : 812-826