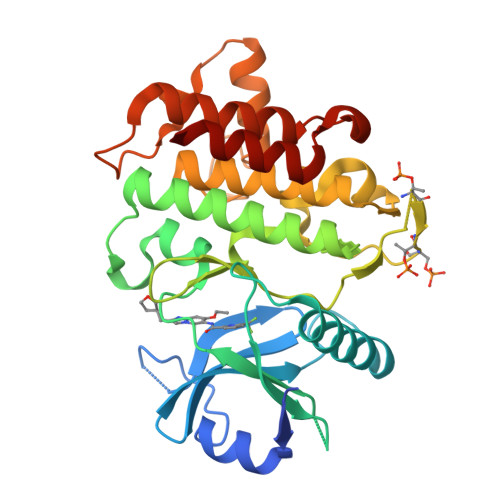
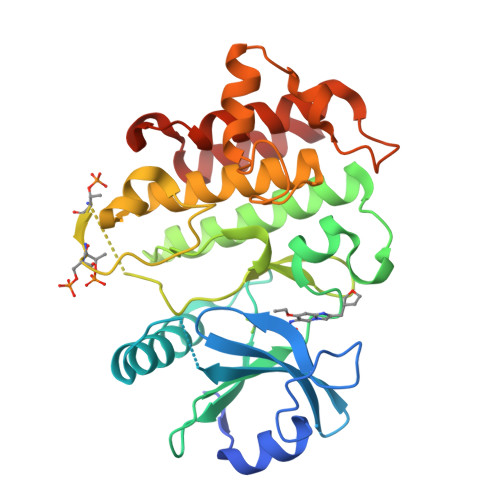
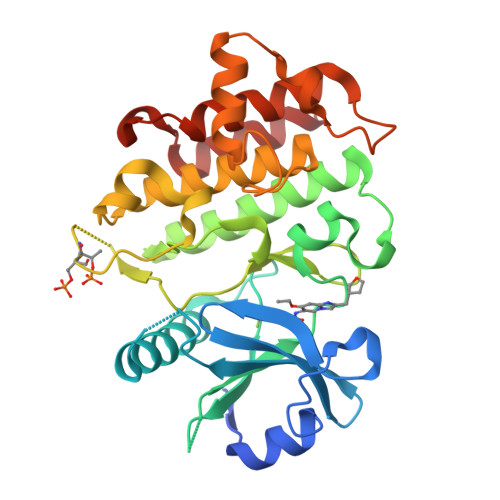
The Discovery of 7-Isopropoxy-2-(1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl)- N -(6-methylpyrazolo[1,5- a ]pyrimidin-3-yl)imidazo[1,2- a ]pyrimidine-6-carboxamide (BIO-7488), a Potent, Selective, and CNS-Penetrant IRAK4 Inhibitor for the Treatment of Ischemic Stroke.
Evans, R., Bolduc, P.N., Pfaffenbach, M., Gao, F., May-Dracka, T., Fang, T., Hopkins, B.T., Chodaparambil, J.V., Henry, K.L., Li, P., Metrick, C., Nelson, A., Trapa, P., Thomas, A., Burkly, L., Peterson, E.A.(2024) J Med Chem 67: 4676-4690
- PubMed: 38467640
- DOI: https://doi.org/10.1021/acs.jmedchem.3c02226
- Primary Citation of Related Structures:
8UCB, 8UCC - PubMed Abstract:
Interleukin receptor-associated kinase 4 (IRAK4) is a key node of signaling within the innate immune system that regulates the production of inflammatory cytokines and chemokines. The presence of d amage- a ssociated m olecular patterns (DAMPs) after tissue damage such as stroke or traumatic brain injury (TBI) initiates signaling through the IRAK4 pathway that can lead to a feed-forward inflammatory loop that can ultimately hinder patient recovery. Herein, we describe the first potent, selective, and CNS-penetrant IRAK4 inhibitors for the treatment of neuroinflammation. Lead compounds from the series were evaluated in CNS PK/PD models of inflammation, as well as a mouse model of ischemic stroke. The SAR optimization detailed within culminates in the discovery of BIO-7488, a highly selective and potent IRAK4 inhibitor that is CNS penetrant and has excellent ADME properties.
Organizational Affiliation:
Department of Medicinal Chemistry, Biogen, 225 Binney Street, Cambridge, Massachusetts 02142, United States.