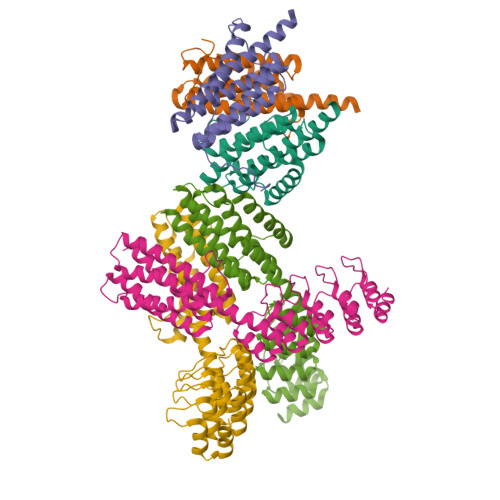
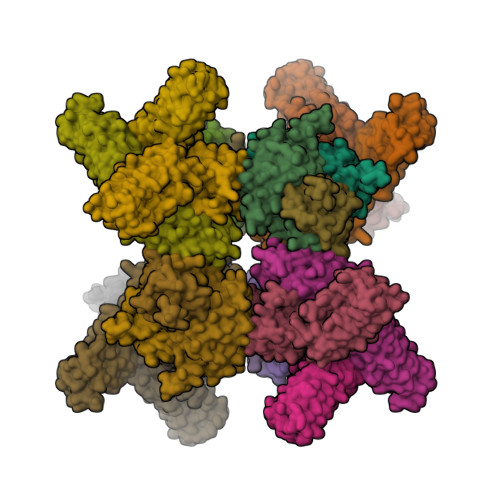
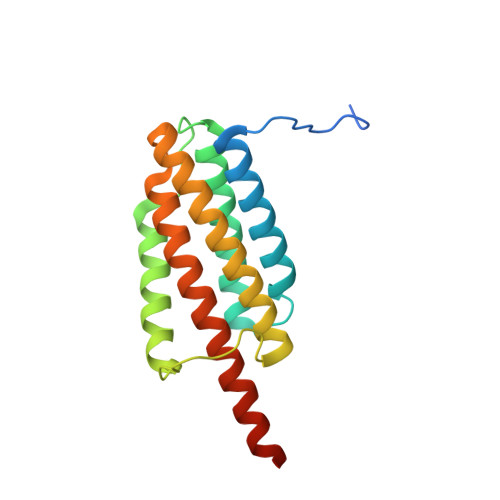
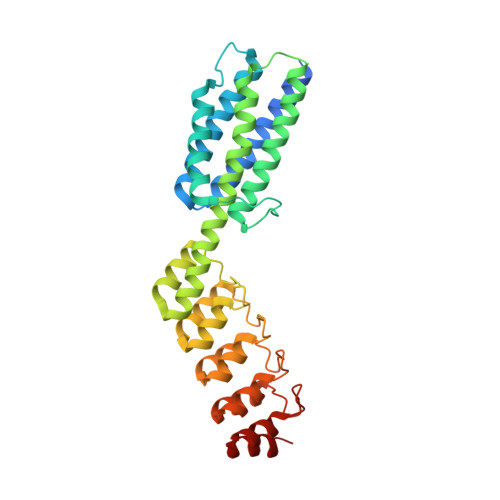
X-ray crystal structure of a designed rigidified imaging scaffold in the ligand-free conformation.
Agdanowski, M.P., Castells-Graells, R., Sawaya, M.R., Cascio, D., Yeates, T.O., Arbing, M.A.(2024) Acta Crystallogr F Struct Biol Commun 80: 107-115
- PubMed: 38767964
- DOI: https://doi.org/10.1107/S2053230X2400414X
- Primary Citation of Related Structures:
8V9O - PubMed Abstract:
Imaging scaffolds composed of designed protein cages fused to designed ankyrin repeat proteins (DARPins) have enabled the structure determination of small proteins by cryogenic electron microscopy (cryo-EM). One particularly well characterized scaffold type is a symmetric tetrahedral assembly composed of 24 subunits, 12 A and 12 B, which has three cargo-binding DARPins positioned on each vertex. Here, the X-ray crystal structure of a representative tetrahedral scaffold in the apo state is reported at 3.8 Å resolution. The X-ray crystal structure complements recent cryo-EM findings on a closely related scaffold, while also suggesting potential utility for crystallographic investigations. As observed in this crystal structure, one of the three DARPins, which serve as modular adaptors for binding diverse `cargo' proteins, present on each of the vertices is oriented towards a large solvent channel. The crystal lattice is unusually porous, suggesting that it may be possible to soak crystals of the scaffold with small (≤30 kDa) protein cargo ligands and subsequently determine cage-cargo structures via X-ray crystallography. The results suggest the possibility that cryo-EM scaffolds may be repurposed for structure determination by X-ray crystallography, thus extending the utility of electron-microscopy scaffold designs for alternative structural biology applications.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California Los Angeles, Los Angeles, CA 90095, USA.