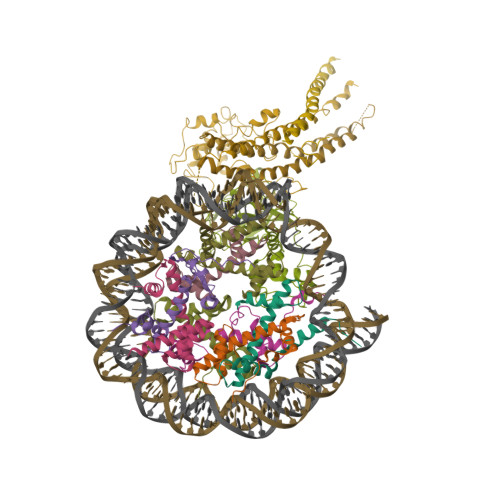
Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Wang, L., Zhang, H., Jia, Q., Li, W., Yang, C., Ma, L., Li, M., Lu, Y., Zhu, H., Zhu, P.(2025) Proc Natl Acad Sci U S A 122: e2414490122-e2414490122
- PubMed: 40100634
- DOI: https://doi.org/10.1073/pnas.2414490122
- Primary Citation of Related Structures:
8X2X, 8X2Y, 8X2Z, 8X30, 8X31, 8X32 - PubMed Abstract:
NuA4 is the only essential acetyltransferase in yeast that can catalyze the acetylation of the histones H2A, H2A.Z, and H4, thereby affecting gene transcription. However, the acetylation process of NuA4, such as how NuA4 acetylates H4 and H2A.Z differently, remains largely elusive. Here, using cryoelectron microscopy (cryo-EM) single particle analysis, we present seven cryo-EM structures of piccolo NuA4 (pNuA4) in complex with wild-type H2A.Z or H2A.Z-mutant-containing nucleosomes in the absence or presence of acetyl coenzyme A (Ac-CoA). We revealed that, in the absence of Ac-CoA, pNuA4 adopts multiple conformations to search for its substrates. After adding Ac-CoA, the single-molecule Förster resonance energy transfer (smFRET) and cryo-EM data indicated that pNuA4 prefers to bind H4 and undergoes a dynamic conformational change to complete the acetylation. We also obtained previously unseen structures in states associated with the acetylation of H2A.Z. These cryo-EM structures and smFRET results suggest a complex acetylation process on H4 and H2A.Z by pNuA4. The results provide a comprehensive picture of the mechanism by which pNuA4 acetylates its substrates within an H2A.Z-containing nucleosome.
Organizational Affiliation:
Key Laboratory of Epigenetic Regulation and Intervention, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.