Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Zhou, F., Zhang, Y., Zhu, Y., Zhou, Q., Shi, Y., Hu, Q.(2024) Elife
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial | 725 | Homo sapiens | Mutation(s): 0 EC: 6.4.1.4 | 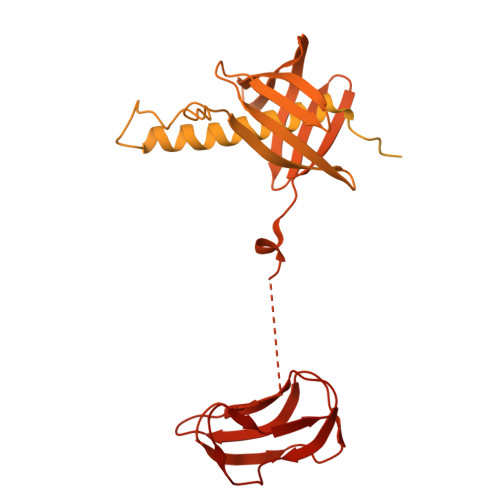 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96RQ3 (Homo sapiens) Explore Q96RQ3 Go to UniProtKB: Q96RQ3 | |||||
PHAROS: Q96RQ3 GTEx: ENSG00000078070 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96RQ3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | 563 | Homo sapiens | Mutation(s): 0 EC: 6.4.1.4 | 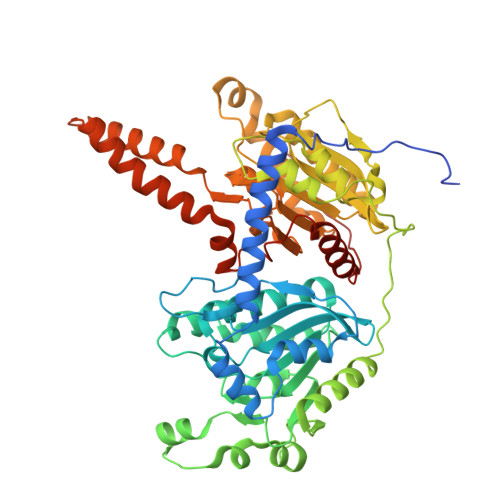 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9HCC0 (Homo sapiens) Explore Q9HCC0 Go to UniProtKB: Q9HCC0 | |||||
PHAROS: Q9HCC0 GTEx: ENSG00000131844 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9HCC0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ACO (Subject of Investigation/LOI) Query on ACO | N [auth B] O [auth B] R [auth F] U [auth J] V [auth J] | ACETYL COENZYME *A C23 H38 N7 O17 P3 S ZSLZBFCDCINBPY-ZSJPKINUSA-N |  | ||
| BTN (Subject of Investigation/LOI) Query on BTN | M [auth A] P [auth C] Q [auth E] S [auth G] T [auth I] | BIOTIN C10 H16 N2 O3 S YBJHBAHKTGYVGT-ZKWXMUAHSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |