Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
Wu, D., Yin, R., Chen, G., Ribeiro-Filho, H.V., Cheung, M., Robbins, P.F., Mariuzza, R.A., Pierce, B.G.(2024) Sci Adv 10: eadq6150-eadq6150
- PubMed: 39576860
- DOI: https://doi.org/10.1126/sciadv.adq6150
- Primary Citation of Related Structures:
8YIV, 8YJ2, 8YJ3 - PubMed Abstract:
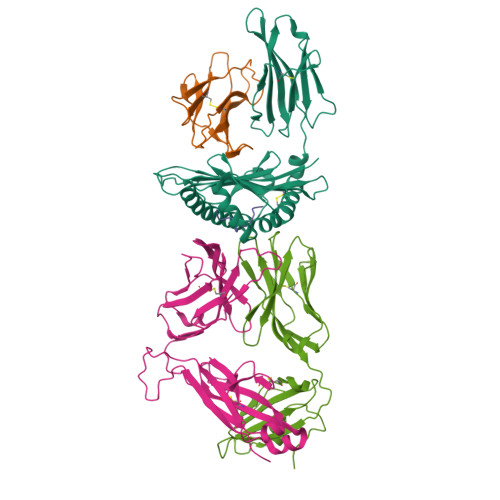
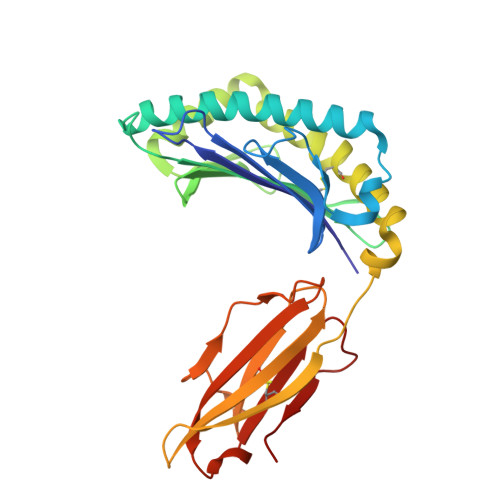
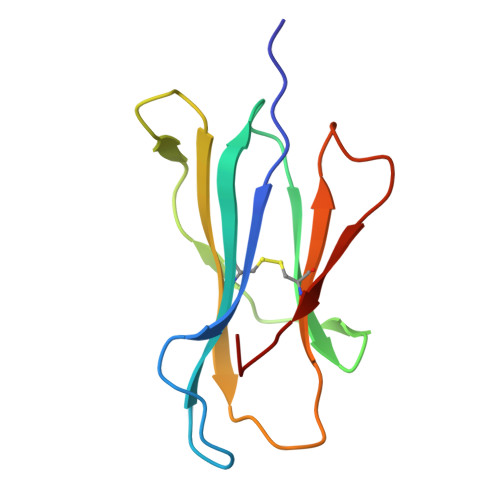
T cell receptors (TCRs) that recognize cancer neoantigens are important for anticancer immune responses and immunotherapy. Understanding the structural basis of TCR recognition of neoantigens provides insights into their exquisite specificity and can enable design of optimized TCRs. We determined crystal structures of a human TCR in complex with NRAS Q61K and Q61R neoantigen peptides and HLA-A1 major histocompatibility complex (MHC), revealing the molecular underpinnings for dual recognition and specificity versus wild-type NRAS peptide. We then used multiple versions of AlphaFold to model the corresponding complex structures, given the challenge of immune recognition for such methods. One implementation of AlphaFold2 (TCRmodel2) with additional sampling was able to generate accurate models of the complexes, while AlphaFold3 also showed strong performance, although success was lower for other complexes. This study provides insights into TCR recognition of a shared cancer neoantigen as well as the utility and practical considerations for using AlphaFold to model TCR-peptide-MHC complexes.
Organizational Affiliation:
Department of Hepatopancreatobiliary Surgery, The First Affiliated Hospital, Laboratory of Structural Immunology, Hengyang Medical School, University of South China, Hengyang, Hunan 421001, China.