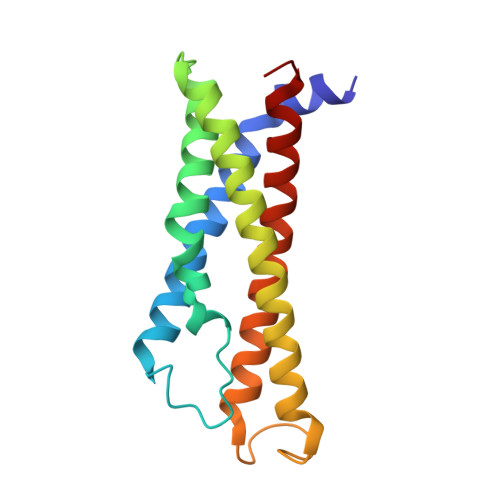
Molecular mechanism of IgE-mediated Fc epsilon RI activation.
Chen, M., Su, Q., Shi, Y.(2024) Nature
- PubMed: 39442557
- DOI: https://doi.org/10.1038/s41586-024-08229-8
- Primary Citation of Related Structures:
8YVU, 8YWA - PubMed Abstract:
Allergic diseases, affecting over a quarter of individuals in industrialized countries, have become significant public health concerns 1,2 . The high-affinity Fc receptor for IgE (FcεRI), mainly present on mast cells and basophils, plays a crucial role in allergic diseases 3-5 . Monomeric IgE binding to FcεRI regulates mast cell survival, differentiation, and maturation 6-8 . However, the underlying molecular mechanism remains unclear. Here we demonstrate that, prior to IgE binding, FcεRI mostly exists as a homo-dimer on human mast cell membrane. The structure of human FcεRI confirms the dimeric organization, with each promoter comprising one α subunit, one β subunit, and two γ subunits. The transmembrane helices of the α subunits form a layered arrangement with those of the γ and β subunits. The dimeric interface is mediated by a four-helix bundle of the α and γ subunits at the intracellular juxtamembrane region. Cholesterol-like molecules embedded within the transmembrane domain may stabilize the dimeric assembly. Upon IgE binding, the dimeric FcεRI dissociates into two protomers, each binding to an IgE molecule. Importantly, this process elicits transcriptional activation of Egr1/3 and Ccl2 in rat basophils, which can be attenuated by inhibiting the FcεRI dimer-to-monomer transition. Collectively, our study unveils the mechanism of antigen-independent, IgE-mediated FcεRI activation.
Organizational Affiliation:
Research Center for Industries of the Future, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Institute of Biology, Westlake Institute for Advanced Study, Xihu District, Hangzhou, Zhejiang Province, China.