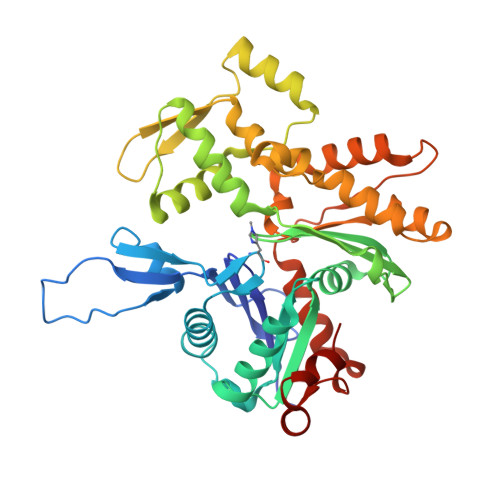
Molecular mechanisms linking missense ACTG2 mutations to visceral myopathy.
Ceron, R.H., Baez-Cruz, F.A., Palmer, N.J., Carman, P.J., Boczkowska, M., Heuckeroth, R.O., Ostap, E.M., Dominguez, R.(2024) Sci Adv 10: eadn6615-eadn6615
- PubMed: 38820162
- DOI: https://doi.org/10.1126/sciadv.adn6615
- Primary Citation of Related Structures:
8V2O, 8V30 - PubMed Abstract:
Visceral myopathy is a life-threatening disease characterized by muscle weakness in the bowel, bladder, and uterus. Mutations in smooth muscle γ-actin (ACTG2) are the most common cause of the disease, but the mechanisms by which the mutations alter muscle function are unknown. Here, we examined four prevalent ACTG2 mutations (R40C, R148C, R178C, and R257C) that cause different disease severity and are spread throughout the actin fold. R178C displayed premature degradation, R148C disrupted interactions with actin-binding proteins, R40C inhibited polymerization, and R257C destabilized filaments. Because these mutations are heterozygous, we also analyzed 50/50 mixtures with wild-type (WT) ACTG2. The WT/R40C mixture impaired filament nucleation by leiomodin 1, and WT/R257C produced filaments that were easily fragmented by smooth muscle myosin. Smooth muscle tropomyosin isoform Tpm1.4 partially rescued the defects of R40C and R257C. Cryo-electron microscopy structures of filaments formed by R40C and R257C revealed disrupted intersubunit contacts. The biochemical and structural properties of the mutants correlate with their genotype-specific disease severity.
Organizational Affiliation:
Department of Physiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.