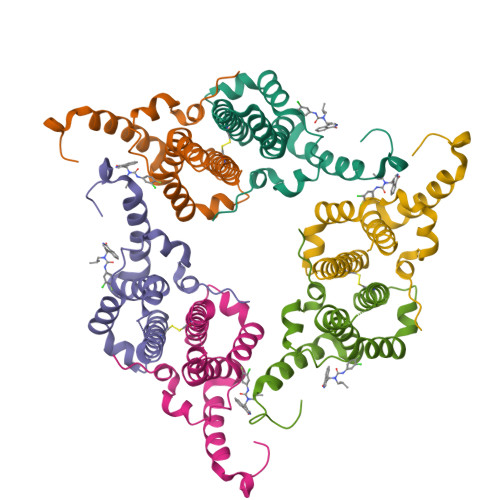
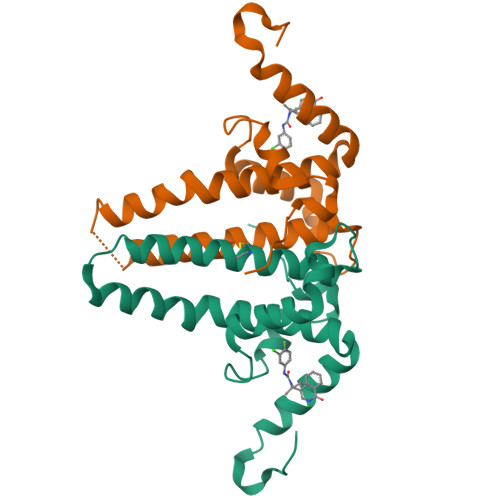
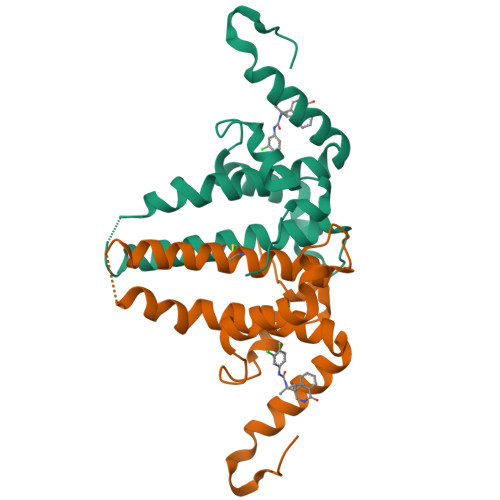
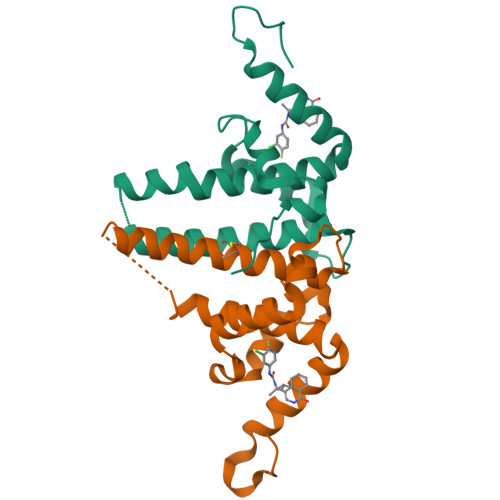
Rational Design, Synthesis, and Structure-Activity Relationship of a Novel Isoquinolinone-Based Series of HBV Capsid Assembly Modulators Leading to the Identification of Clinical Candidate AB-836.
Cole, A.G., Kultgen, S.G., Mani, N., Fan, K.Y., Ardzinski, A., Stever, K., Dorsey, B.D., Mesaros, E.F., Thi, E.P., Graves, I., Tang, S., Harasym, T.O., Lee, A.C.H., Olland, A., Suto, R.K., Sofia, M.J.(2024) J Med Chem 67: 16773-16795
- PubMed: 39231272
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01568
- Primary Citation of Related Structures:
9C9V - PubMed Abstract:
Inhibition of Hepatitis B Virus (HBV) replication by small molecules that modulate capsid assembly and the encapsidation of pgRNA and viral polymerase by HBV core protein is a clinically validated approach toward the development of new antivirals. Through definition of a minimal pharmacophore, a series of isoquinolinone-based capsid assembly modulators (CAMs) was identified. Structural biology analysis revealed that lead molecules possess a unique binding mode, exploiting electrostatic interactions with accessible phenylalanine and tyrosine residues. Key analogs demonstrated excellent primary potency, absorption, distribution, metabolism, and excretion (ADME) and pharmacokinetic properties, and efficacy in a mouse model of HBV. The optimized lead also displayed potent inhibition of capsid uncoating in HBV-infected HepG2 cells expressing the sodium-taurocholate cotransporting polypeptide (NTCP) receptor, affecting the generation of HBsAg and cccDNA establishment. Based on these results, isoquinolinone derivative AB-836 was advanced into clinical development. In Phase 1b trials, AB-836 demonstrated >3 log 10 reduction in serum HBV DNA, however, further development was discontinued due to the observation of incidental alanine aminotransferase (ALT) elevations.
Organizational Affiliation:
Arbutus Biopharma, Inc., 701 Veterans Circle, Warminster, Pennsylvania 18974, United States.