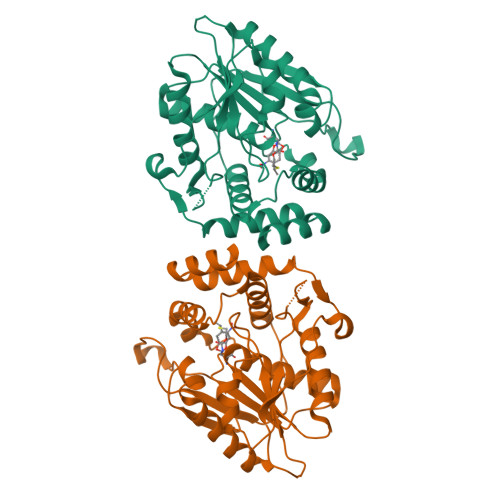
Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation.
McCullough, T.M., Choudhary, V., Akey, D.L., Skiba, M.A., Bernard, S.M., Kittendorf, J.D., Schmidt, J.J., Sherman, D.H., Smith, J.L.(2024) ACS Catal 14: 12551-12563
- PubMed: 39927088
- DOI: https://doi.org/10.1021/acscatal.4c03637
- Primary Citation of Related Structures:
9CBD, 9CEL, 9CFJ, 9CGL, 9CGN, 9CGO - PubMed Abstract:
Emerging antibiotic resistance requires continual improvement in the arsenal of antimicrobial drugs, especially the critical macrolide antibiotics. Formation of the macrolactone scaffold of these polyketide natural products is catalyzed by a modular polyketide synthase (PKS) thioesterase (TE). The TE accepts a linear polyketide substrate from the termina PKS acyl carrier protein to generate an acyl-enzyme adduct that is resolved by attack of a substrate hydroxyl group to form the macrolactone. Our limited mechanistic understanding of TE selectivity for a substrate nucleophile and/or water has hampered development of TEs as biocatalysts that accommodate a variety of natural and non-natural substrates. To understand how TEs direct the substrate nucleophile for macrolactone formation, acyl-enzyme intermediates were trapped as stable amides by substituting the natural serine OH with an amino group. Incorporation of the unnatural amino acid, 1,3-diaminopropionic acid (DAP), was tested with five PKS TEs. DAP-modified TEs (TE DAP ) from the pikromycin and erythromycin pathways were purified and tested with six full-length polyketide intermediates from three pathways. The erythromycin TE had permissive substrate selectivity, whereas the pikromycin TE was selective for its native hexaketide and heptaketide substrates. In a crystal structure of a native substrate trapped in pikromycin TE DAP , the linear heptaketide was curled in the active site with the nucleophilic hydroxyl group positioned 4 Å from the amide-enzyme linkage. The curled heptaketide displayed remarkable shape complementarity with the TE acyl cavity. The strikingly different shapes of acyl cavities in TEs of known structure, including those reported here for juvenimicin, tylosin and fluvirucin biosynthesis, provide insights to facilitate TE engineering and optimization.
Organizational Affiliation:
Life Sciences Institute, Mary Sue Coleman Hall, 210 Washtenaw Ave., University of Michigan, Ann Arbor, MI 48109-2216, United States.