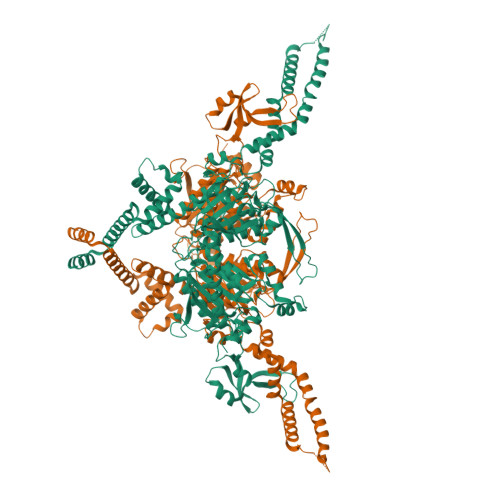
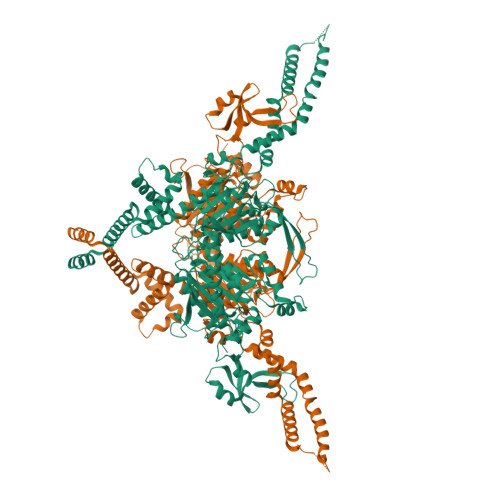
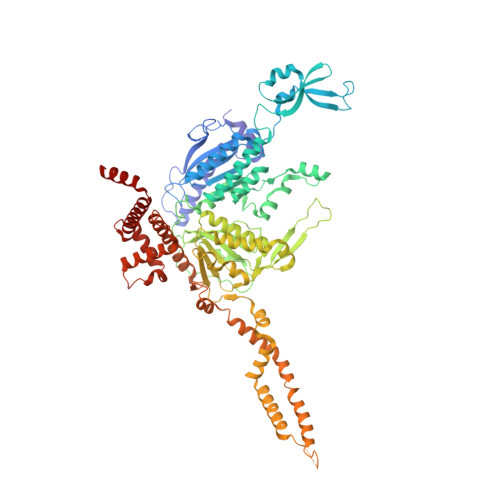
Structure of the endogenous insect acetyl-coA carboxylase carboxyltransferase domain.
Wang, D., Bu, F., Yang, G., Brenke, H., Liu, B.(2024) J Biological Chem 300: 107800-107800
- PubMed: 39305960
- DOI: https://doi.org/10.1016/j.jbc.2024.107800
- Primary Citation of Related Structures:
9CV6 - PubMed Abstract:
Acetyl-coenzyme A carboxylases (ACCs) are pivotal in fatty acid metabolism, converting acetyl-CoA to malonyl-CoA. While ACCs in humans, plants, and microbes have been extensively studied, insect ACCs, crucial for lipid biosynthesis and physiological processes, remain relatively unexplored. Unlike mammals, which have ACC1 and ACC2 in different tissues, insects possess a single ACC gene, underscoring its unique role in their metabolism. Noctuid moths, such as Trichoplusia ni, are major agricultural pests causing significant crop damage and economic loss. Their resistance to both biological and synthetic insecticides complicates pest control. Recent research has introduced cyclic ketoenols as novel insecticides targeting ACCs, yet structural information to guide their design is limited. Here, we present a 3.12 Å cryo-EM structure of the carboxyltransferase (CT) domain of T. ni ACC, offering the first detailed structural insights into insect ACCs. Our structural comparisons with ACC CT domains from other species and analyses of drug binding sites can guide future drug modification and design. Notably, unique interactions between the CT and the central domain in T. ni ACC provide new directions for studying the ACC holoenzyme. These findings contribute valuable information for pest control and basic biological understanding of lipid biosynthesis.
Organizational Affiliation:
The Hormel Institute, University of Minnesota, Austin, MN, USA.