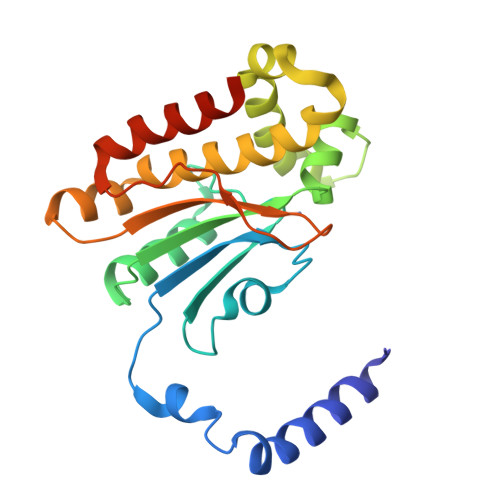
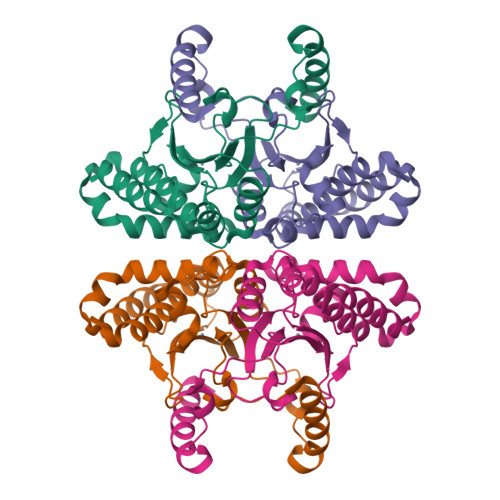
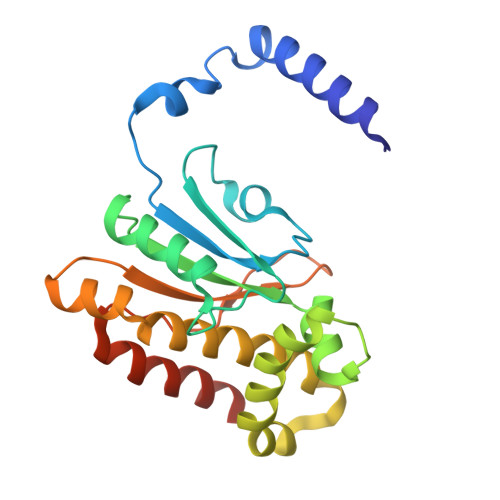
Serendipitous high-resolution structure of Escherichia coli carbonic anhydrase 2.
Rankin, M.R., Smith, J.L.(2025) Acta Crystallogr F Struct Biol Commun
- PubMed: 39812168
- DOI: https://doi.org/10.1107/S2053230X25000068
- Primary Citation of Related Structures:
9EAT, 9EAW, 9EBZ - PubMed Abstract:
X-ray crystallography remains the dominant method of determining the three-dimensional structure of proteins. Nevertheless, this resource-intensive process may be hindered by the unintended crystallization of contaminant proteins from the expression source. Here, the serendipitous discovery of two novel crystal forms and one new, high-resolution structure of carbonic anhydrase 2 (CA2) from Escherichia coli that arose during a crystallization campaign for an unrelated target is reported. By comparing unit-cell parameters with those in the PDB, contaminants such as CA2 can be identified, preventing futile molecular-replacement attempts. Crystallographers can use these new lattice parameters to diagnose CA2 contamination in similar experiments.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, Ann Arbor, MI 48109, USA.