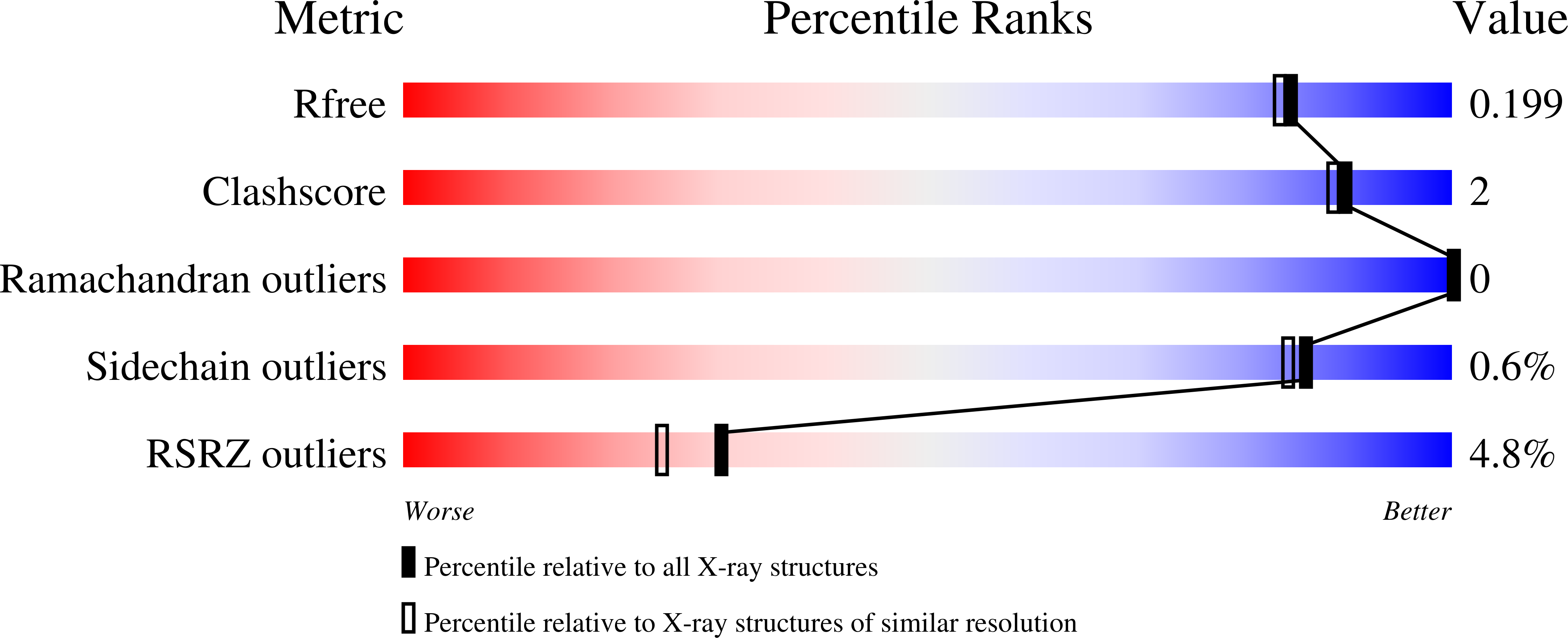
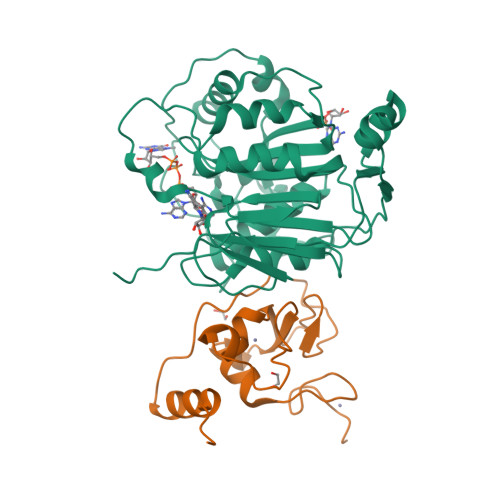
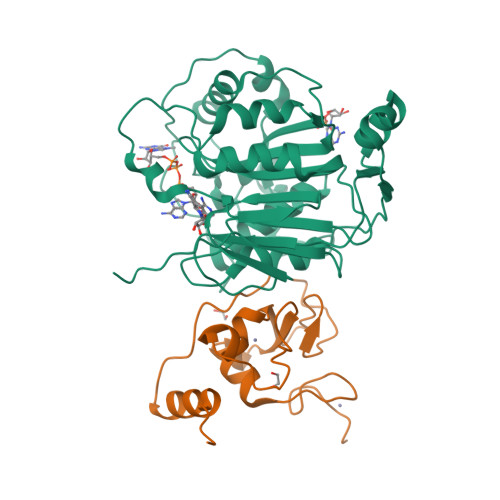
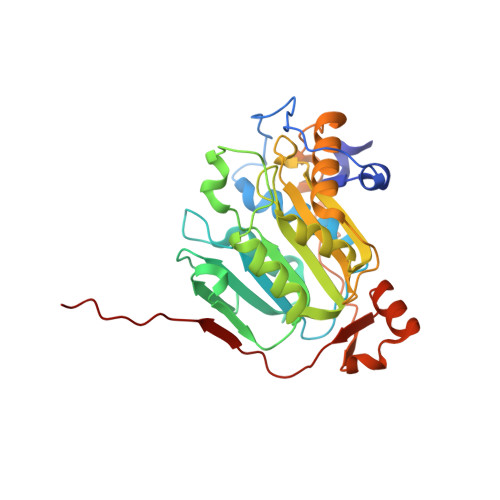
SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
Kremling, V., Sprenger, J., Oberthuer, D., Chapman, H.N., Middendorf, P., Fernandez-Garcia, Y., Ehrt, C., Falke, S., Kiene, A., Klopprogge, B., Scheer, T.E.S., Rarey, M., Barthels, F., Kuehn, S., Guenther, S.To be published.