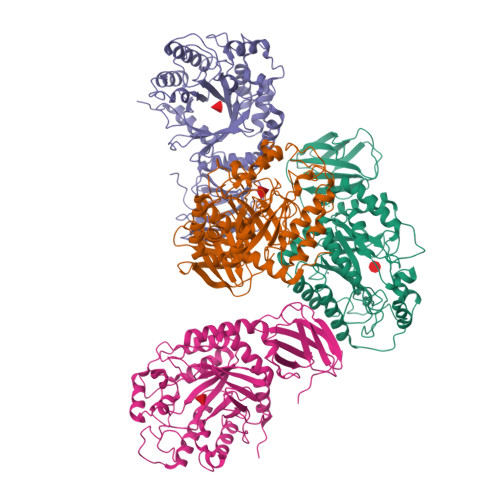
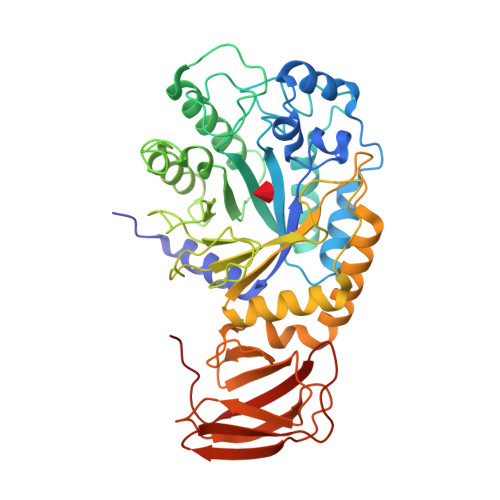
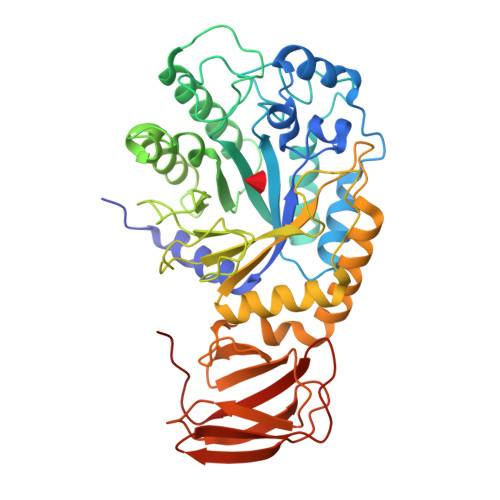
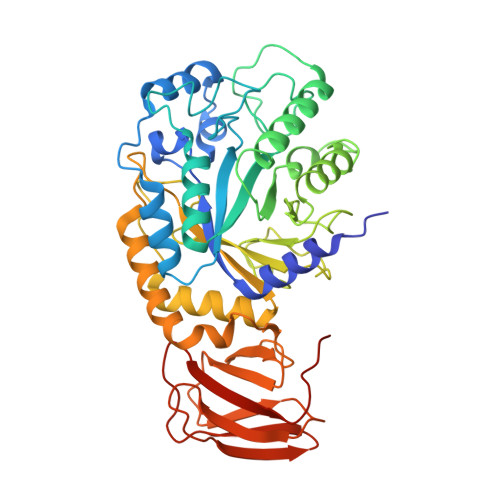
Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Yang, Y., Holck, J., Thorhallsson, A.T., Hunt, C.J., Yang, H., Morth, J.P., Meyer, A.S., Zeuner, B.(2025) FEBS J 292: 653-680
- PubMed: 39658312
- DOI: https://doi.org/10.1111/febs.17347
- Primary Citation of Related Structures:
9EU1, 9EU2, 9EU3, 9EU4 - PubMed Abstract:
GH29A α-l-fucosidases (EC 3.2.1.51) catalyze the release of α-l-fucosyl moieties from the nonreducing end of glycoconjugates by hydrolysis and some also catalyze transglycosylation. The latter is particularly interesting with regard to designing enzymatic synthesis of human milk oligosaccharides (HMOs). We combined the bioinformatics tool conserved unique peptide patterns (CUPP) and phylogenetic clustering to discover new microbial GH29A α-l-fucosidases of the underexplored CUPP group GH29:13.1. Three uncharacterized bacterial enzymes (EaGH29, SeGH29, and PmGH29) and two previously identified GH29A α-l-fucosidases (BF3242 and TfFuc1) were selected for reaction optimization, biochemical, and structural characterization. Kinetics, pH-temperature optima, and substrate preference for 2-chloro-4-nitrophenyl-α-l-fucopyranoside (CNP-α-l-Fuc) and 2'-fucosyllactose (2'FL) were determined. Transglycosylation was favored at high neutral to alkaline pH, especially for EaGH29, SeGH29, TfFuc1, and BF3242, mainly because hydrolysis was decreased. The α-l-fucosidases exhibited medium regioselectivity in transglycosylation, generally forming two out of five detected lacto-N-fucopentaose (LNFP) isomers from 2'FL and lacto-N-tetraose (LNT). Alkaline pH also affected the transglycosylation product regioselectivity of SeGH29, which was also affected by a Leu/Phe exchange in the acceptor binding site. New crystal structures of TfFuc1 and BF3242 showed congruence in active site topology between these two enzymes and contributed to understanding the function of GH29A α-l-fucosidases. Notably, the structural data provide new insight into the role of an Asn residue located between the two catalytic residues in the active site.
Organizational Affiliation:
School of Food and Biological Engineering, Jiangsu University, Zhenjiang, China.