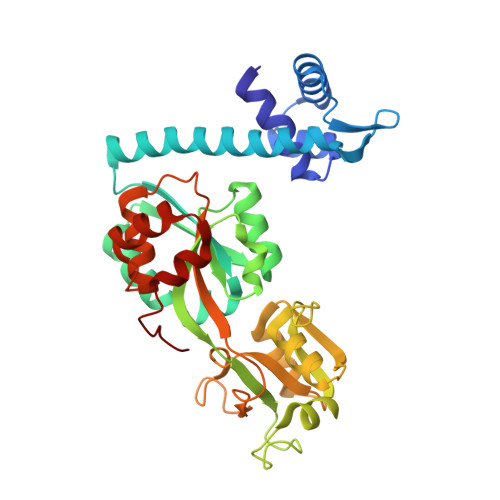
The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Verschueren, K.H.G., Dodson, E.J., Wilkinson, A.J.(2024) Eur Biophys J 53: 311-326
- PubMed: 38976018
- DOI: https://doi.org/10.1007/s00249-024-01716-w
- Primary Citation of Related Structures:
9F14, 9FDD - PubMed Abstract:
In Escherichia coli and Salmonella typhimurium, cysteine biosynthesis requires the products of 20 or more cys genes co-ordinately regulated by CysB. Under conditions of sulphur limitation and in the presence of the inducer, N-acetylserine, CysB binds to cys promoters and activates the transcription of the downstream coding sequences. CysB is a homotetramer, comprising an N-terminal DNA binding domain (DBD) and a C-terminal effector binding domain (EBD). The crystal structure of a dimeric EBD fragment of CysB from Klebsiella aerogenes revealed a protein fold similar to that seen in Lac repressor but with a different symmetry in the dimer so that the mode of DNA binding was not apparent. To elucidate the subunit arrangement in the tetramer, we determined the crystal structure of intact CysB in complex with N-acetylserine. The tetramer has two subunit types that differ in the juxtaposition of their winged helix-turn-helix DNA binding domains with respect to the effector binding domain. In the assembly, the four EBDs form a core with the DNA binding domains arranged in pairs on the surface. N-acetylserine makes extensive polar interactions in an enclosed binding site, and its binding is accompanied by substantial conformational rearrangements of surrounding residues that are propagated to the protein surface where they appear to alter the arrangement of the DNA binding domains. The results are (i) discussed in relation to the extensive mutational data available for CysB and (ii) used to propose a structural mechanism of N-acetylserine induced CysB activation.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, University of York, York, YO10 5DD, UK.