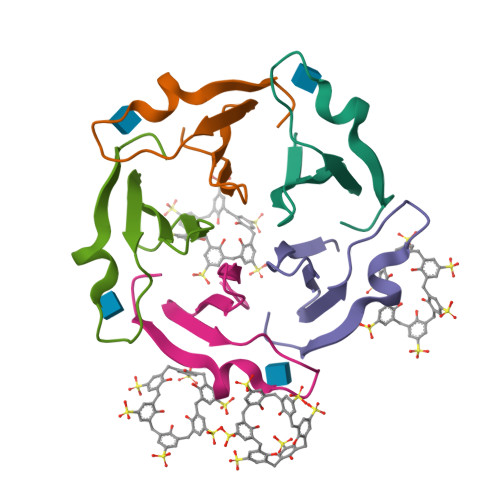
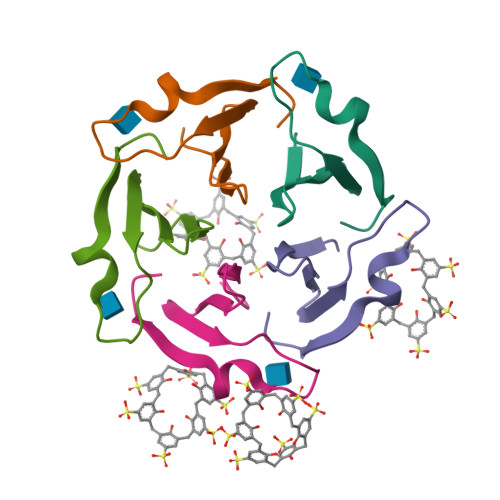
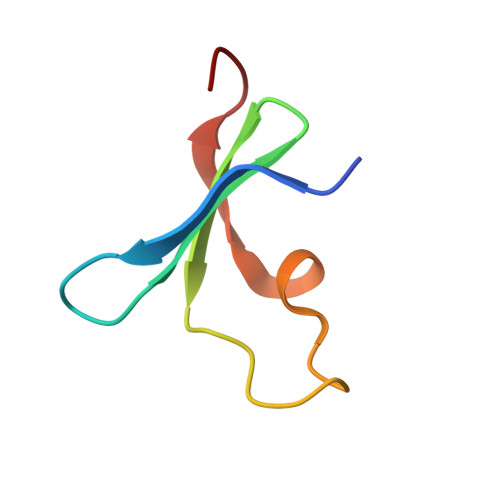
A Macrocycle-Mediated Protein Cage.
Flood, R.J., Thureau, A., Crowley, P.B.(2024) ACS Macro Lett 13: 1686-1690
- PubMed: 39592260
- DOI: https://doi.org/10.1021/acsmacrolett.4c00656
- Primary Citation of Related Structures:
9FRO - PubMed Abstract:
Engineered protein cages are of great interest considering their diverse applications in delivery and catalysis. Here, we describe macrocycle-triggered icosahedral cage assembly of a designed β-propeller. Cage assembly was evidenced by small-angle X-ray scattering and X-ray crystallography.
Organizational Affiliation:
SSPC, Science Foundation Ireland Research Centre for Pharmaceuticals, School of Biological and Chemical Sciences, University of Galway, University Road, Galway H91 TK33, Ireland.