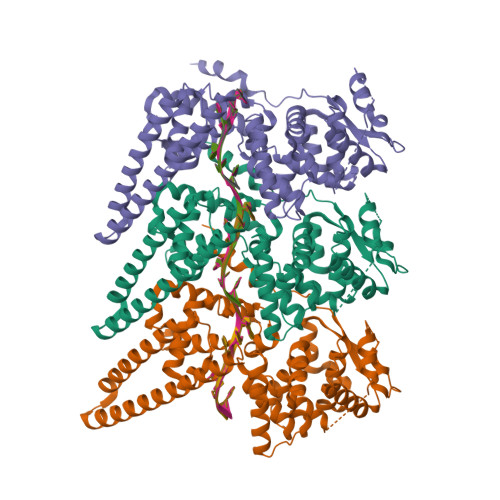
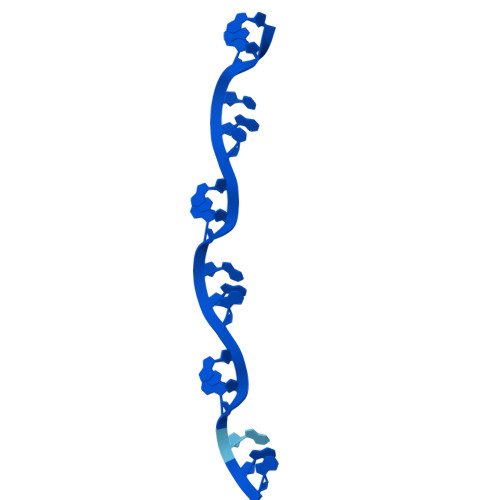
Cryo-EM structure of single-layered nucleoprotein-RNA complex from Marburg virus.
Zinzula, L., Beck, F., Camasta, M., Bohn, S., Liu, C., Morado, D., Bracher, A., Plitzko, J.M., Baumeister, W.(2024) Nat Commun 15: 10307-10307
- PubMed: 39604358
- DOI: https://doi.org/10.1038/s41467-024-54431-7
- Primary Citation of Related Structures:
9FVD - PubMed Abstract:
Marburg virus (MARV) causes lethal hemorrhagic fever in humans, posing a threat to global health. We determined by cryogenic electron microscopy (cryo-EM) the MARV helical ribonucleoprotein (RNP) complex structure in single-layered conformation, which differs from the previously reported structure of a double-layered helix. Our findings illuminate novel RNP interactions and expand knowledge on MARV genome packaging and nucleocapsid assembly, both processes representing attractive targets for the development of antiviral therapeutics against MARV disease.
Organizational Affiliation:
Max Planck Institute of Biochemistry, Research Group Molecular Structural Biology, Martinsried, Germany. zinzula@biochem.mpg.de.