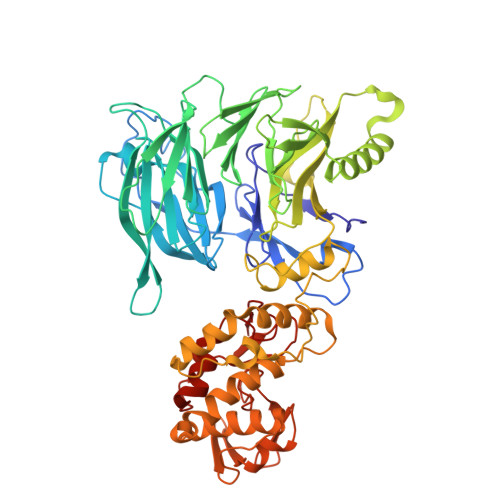
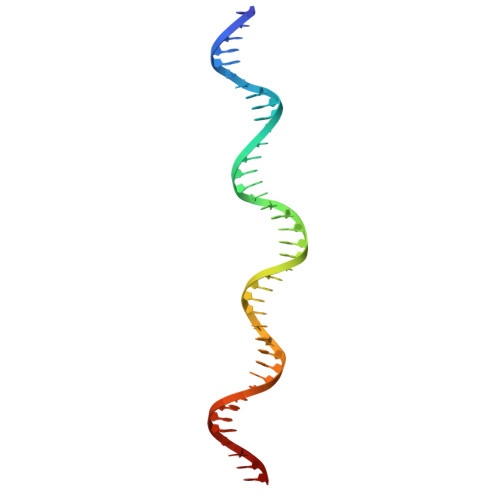
Structural and kinetic insights into tRNA promoter engagement by yeast general transcription factor TFIIIC.
Seifert-Davila, W., Chaban, A., Baudin, F., Girbig, M., Hauptmann, L., Hoffmann, T., Duss, O., Eustermann, S., Muller, C.W.(2025) Nucleic Acids Res 53
- PubMed: 39657784
- DOI: https://doi.org/10.1093/nar/gkae1174
- Primary Citation of Related Structures:
9GC3, 9GCK - PubMed Abstract:
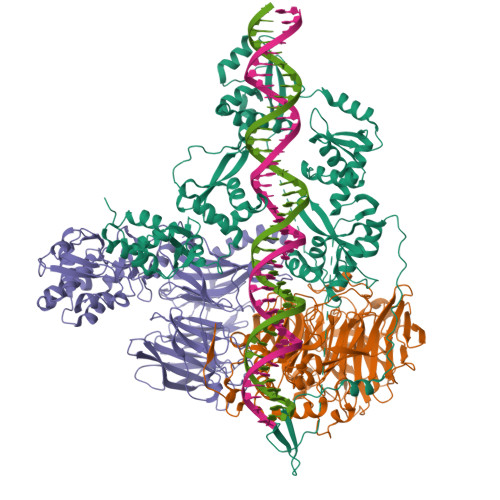
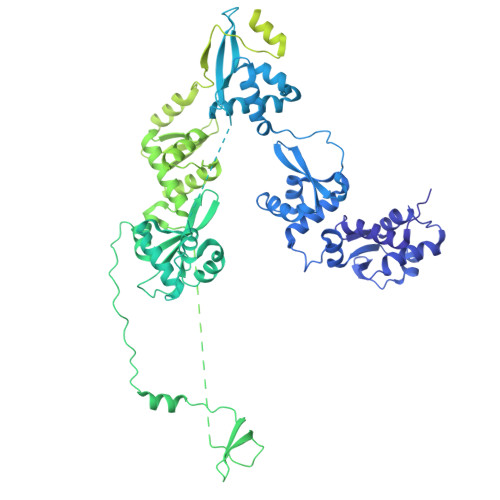
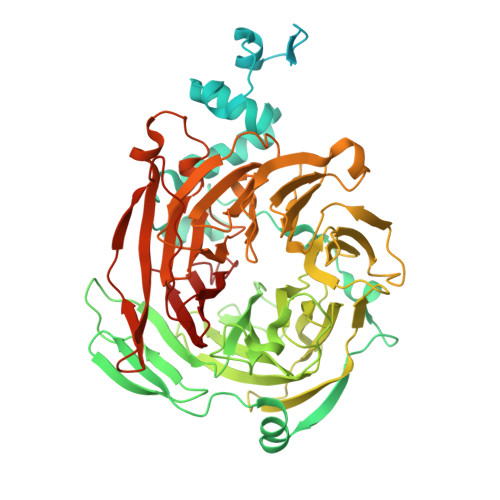
Transcription of transfer RNA (tRNA) genes by RNA polymerase (Pol) III requires the general transcription factor IIIC (TFIIIC), which recognizes intragenic A-box and B-box DNA motifs of type II gene promoters. However, the underlying mechanism has remained elusive, in part due to missing structural information for A-box recognition. In this study, we use single-particle cryogenic electron microscopy (cryo-EM) and single-molecule fluorescence resonance energy transfer (smFRET) to reveal structural and real-time kinetic insights into how the 520-kDa yeast TFIIIC complex engages A-box and B-box DNA motifs in the context of a tRNA gene promoter. Cryo-EM structures of τA and τB subcomplexes bound to the A-box and B-box were obtained at 3.7 and 2.5 Å resolution, respectively, while cryo-EM single-particle mapping determined the specific distance and relative orientation of the τA and τB subcomplexes revealing a fully engaged state of TFIIIC. smFRET experiments show that overall recruitment and residence times of TFIIIC on a tRNA gene are primarily governed by B-box recognition, while footprinting experiments suggest a key role of τA and the A-box in TFIIIB and Pol III recruitment following TFIIIC recognition of type II promoters.
Organizational Affiliation:
Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Meyerhofstrasse 1, 69117 Heidelberg, Germany.