Full Text |
QUERY: Structure Author = "Zang, X." | MyPDB Login | Search API |
| Search Summary | This query matches 13 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry Type | 1 to 13 of 13 Structures Page 1 of 1 Sort by
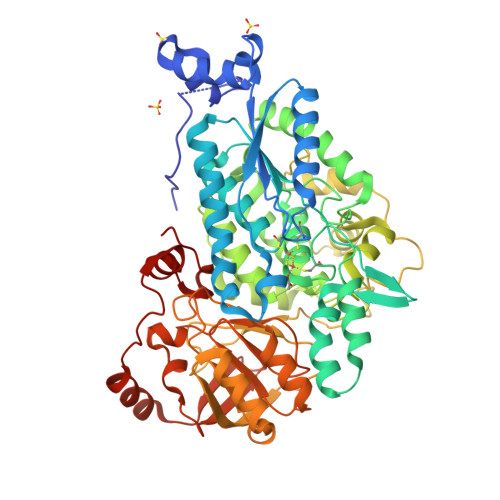
The crystal structure of DHADZang, X., Huang, W.X., Cheng, R., Wu, L., Zhou, J.H., Tang, Y., Yan, Y. To be published
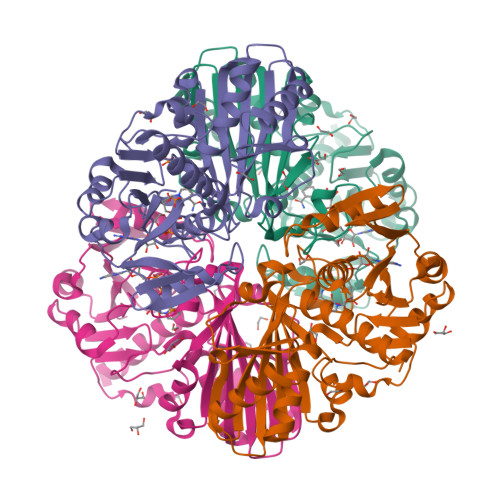
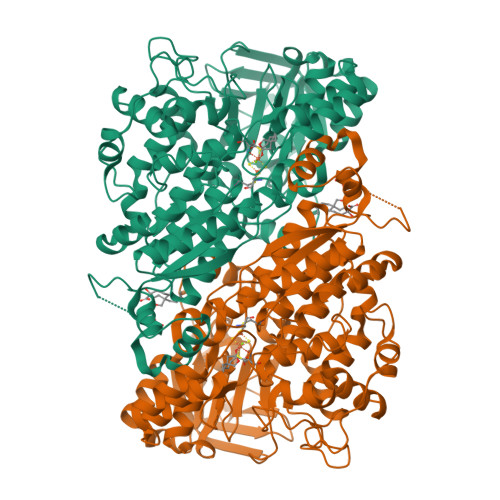
A fungal glyceraldehyde-3-phosphate dehydrogenase with self-resistance to inhibitor heptelidic acidYan, Y., Zang, X., Cooper, S.J., Lin, H., Zhou, J., Tang, Y. (2020) Chem Sci 11: 9554-9562
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with inhibitor heptelidic acidYan, Y., Zang, X., Cooper, S.J., Lin, H., Zhou, J., Tang, Y. (2020) Chem Sci 11: 9554-9562
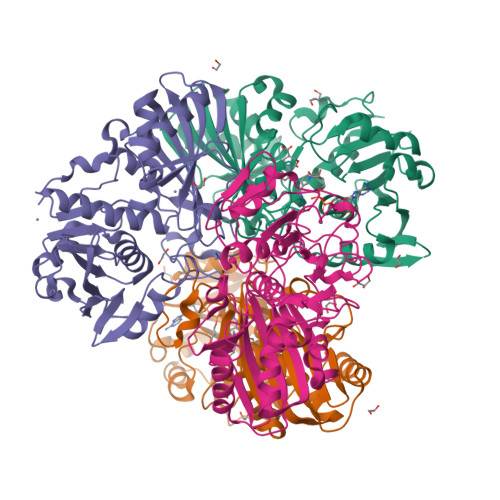
Acetolactate Synthase from Trichoderma harzianum with inhibitor harzianic acidZang, X., Xie, L., Chen, M., Tang, Y., Zhou, J. (2021) J Am Chem Soc
Acetolactate Synthase from Trichoderma harzianum(2021) J Am Chem Soc
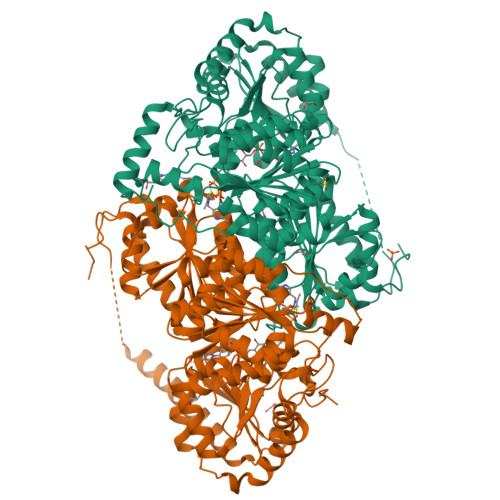
The mutant structure of DHAD V178WZhou, J., Zang, X., Tang, Y., Yan, Y. (2024) Biodes Res 6: 0046-0046
Dihydroxyacid dehydratase (DHAD) mutant-V497FZhou, J., Zang, X., Tang, Y., Yan, Y. (2024) Biodes Res 6: 0046-0046
Complex structure of the azoxy synthase VlmB and its substrateZang, X., Zhou, J.H., Yiling, D., Jingkun, S., Zhijie, Z., Zhuanglin, S., Guiyun, Z. To be published
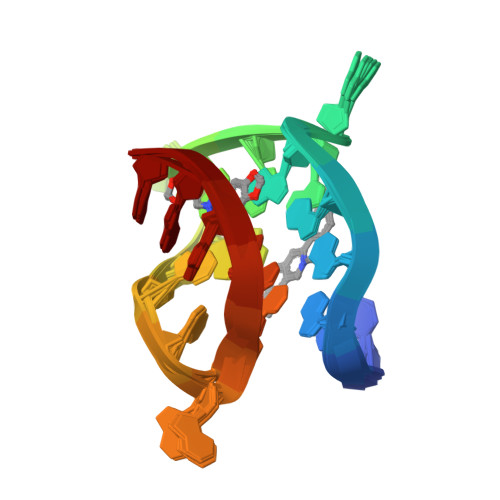
NMR Solution Structure of the 2:1 Coptisine-ATF4-G4 ComplexXiao, C.M., Li, Y.P., Liu, Y.S., Dong, R.F., He, X.Y., Lin, Q., Zang, X., Wang, K.B., Xia, Y.Z., Kong, L.Y. To be published
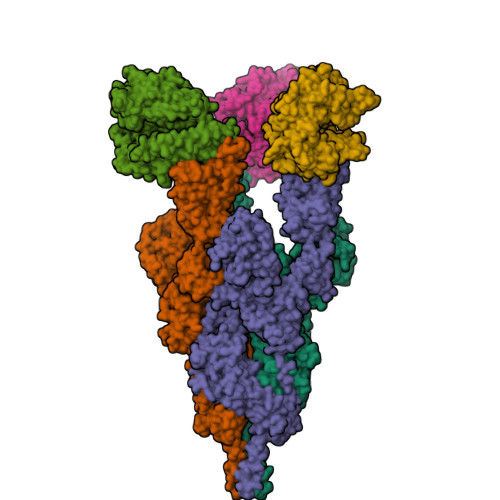
Structure of BA.2.86 spike protein in complex with ACE2.(2024) Nat Commun 15: 8728-8728
1 to 13 of 13 Structures Page 1 of 1 Sort by |