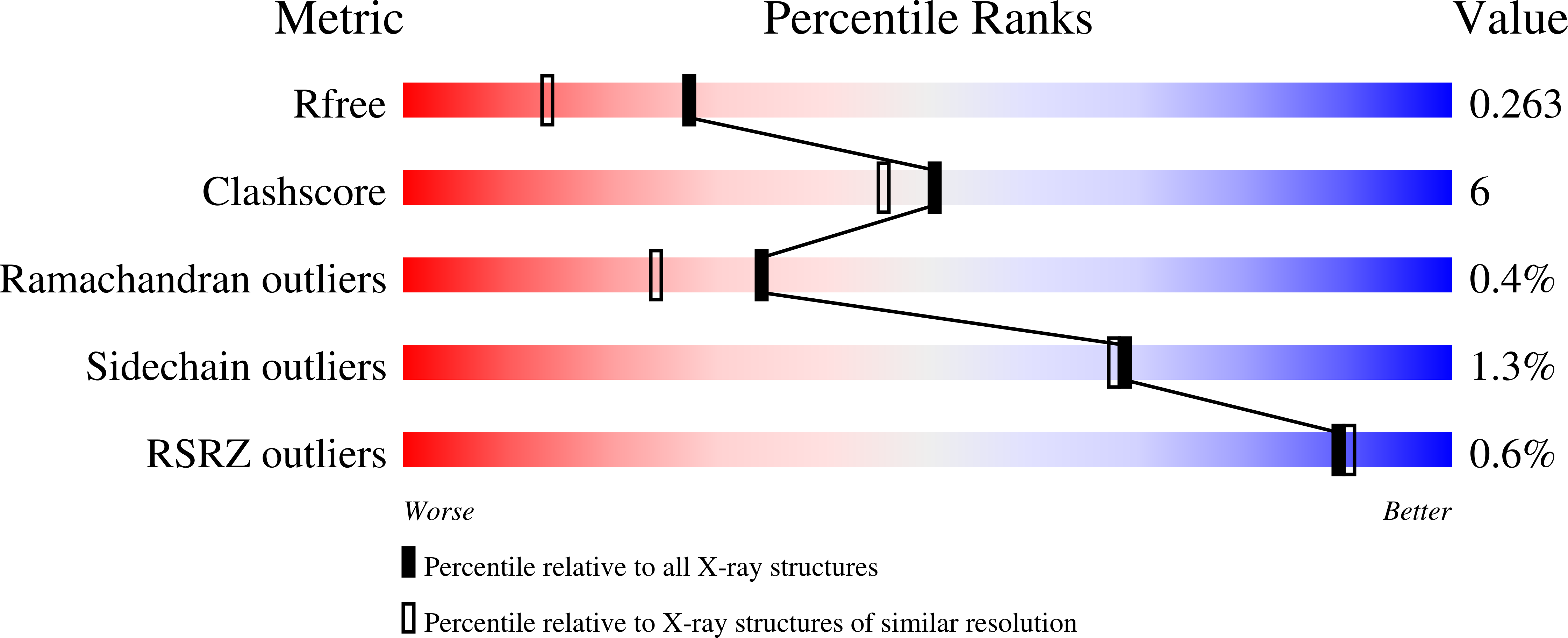
Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Harijan, R.K., Kiema, T.-R., Karjalainen, M.P., Janardan, N., Murthy, M.R.N., Weiss, M.S., Michels, P.A.M., Wierenga, R.K.(2013) Biochem J 455: 119
- PubMed: 23909465
- DOI: https://doi.org/10.1042/BJ20130669
- Primary Citation of Related Structures:
3ZBG, 3ZBK, 3ZBL, 3ZBN, 4BI9, 4BIA - PubMed Abstract:
Thiolases are essential CoA-dependent enzymes in lipid metabolism. In the present study we report the crystal structures of trypanosomal and leishmanial SCP2 (sterol carrier protein, type-2)-thiolases. Trypanosomatidae cause various widespread devastating (sub)-tropical diseases, for which adequate treatment is lacking. The structures reveal the unique geometry of the active site of this poorly characterized subfamily of thiolases. The key catalytic residues of the classical thiolases are two cysteine residues, functioning as a nucleophile and an acid/base respectively. The latter cysteine residue is part of a CxG motif. Interestingly, this cysteine residue is not conserved in SCP2-thiolases. The structural comparisons now show that in SCP2-thiolases the catalytic acid/base is provided by the cysteine residue of the HDCF motif, which is unique for this thiolase subfamily. This HDCF cysteine residue is spatially equivalent to the CxG cysteine residue of classical thiolases. The HDCF cysteine residue is activated for acid/base catalysis by two main chain NH-atoms, instead of two water molecules, as present in the CxG active site. The structural results have been complemented with enzyme activity data, confirming the importance of the HDCF cysteine residue for catalysis. The data obtained suggest that these trypanosomatid SCP2-thiolases are biosynthetic thiolases. These findings provide promise for drug discovery as biosynthetic thiolases catalyse the first step of the sterol biosynthesis pathway that is essential in several of these parasites.
Organizational Affiliation:
*Department of Biochemistry, Biocenter Oulu, University of Oulu, Oulu FIN-90014, Finland.