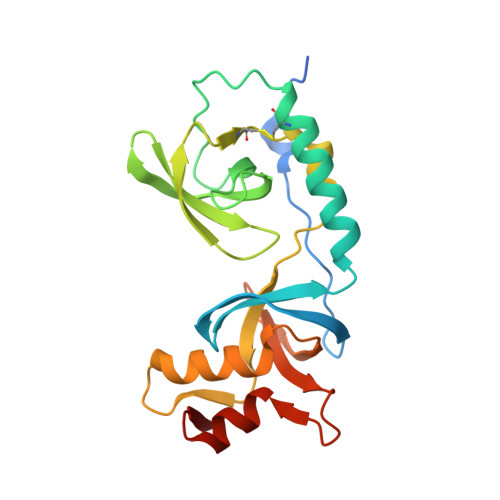
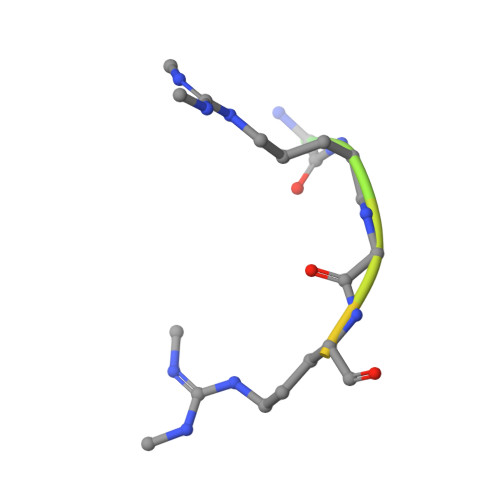
Crystal structure of human TDRD1 extended Tudor domain in complex with a symmetrically dimethylated E2F peptide
Tallant, C., Savitsky, P., Moehlenbrink, J., Chan, C., Nunez-Alonso, G., Newman, J.A., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Fedorov, O., La Thangue, N.B., Knapp, S.To be published.