The crystal structure of plasma-derived human serum albumin
Park, J., Kim, M.S., Shin, D.H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serum albumin | 585 | Homo sapiens | Mutation(s): 0 | 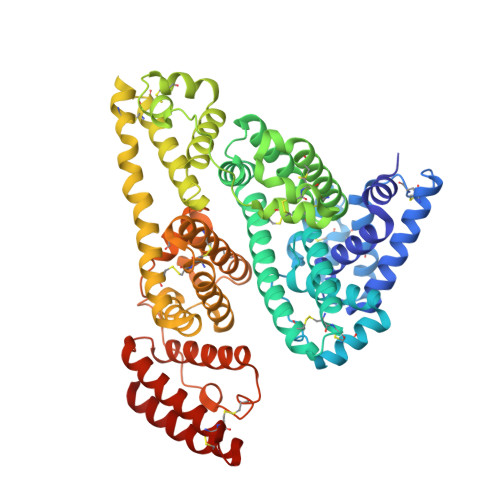 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P02768 (Homo sapiens) Explore P02768 Go to UniProtKB: P02768 | |||||
PHAROS: P02768 GTEx: ENSG00000163631 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EIC (Subject of Investigation/LOI) Query on EIC | D [auth A], TA [auth C], W [auth B] | LINOLEIC ACID C18 H32 O2 OYHQOLUKZRVURQ-HZJYTTRNSA-N |  | ||
| PLM (Subject of Investigation/LOI) Query on PLM | F [auth A], VA [auth C], Y [auth B], Z [auth B] | PALMITIC ACID C16 H32 O2 IPCSVZSSVZVIGE-UHFFFAOYSA-N |  | ||
| 1PE Query on 1PE | RA [auth B] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| TRP (Subject of Investigation/LOI) Query on TRP | AA [auth B], H [auth A], XA [auth C] | TRYPTOPHAN C11 H12 N2 O2 QIVBCDIJIAJPQS-VIFPVBQESA-N |  | ||
| PG4 Query on PG4 | EB [auth C], GB [auth C], NA [auth B], S [auth A], U [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | DB [auth C], KA [auth B], R [auth A], SA [auth B] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| OCA (Subject of Investigation/LOI) Query on OCA | E [auth A], G [auth A], UA [auth C], WA [auth C], X [auth B] | OCTANOIC ACID (CAPRYLIC ACID) C8 H16 O2 WWZKQHOCKIZLMA-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | CB [auth C] FB [auth C] HB [auth C] IA [auth B] JA [auth B] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | AB [auth C] BA [auth B] BB [auth C] CA [auth B] DA [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSO Query on CSO | A, B, C | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 203.576 | α = 90 |
| b = 113.466 | β = 104.878 |
| c = 86.809 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| SCALA | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation (NRF, Korea) | Korea, Republic Of | 2015R1D1A1A01058942 |