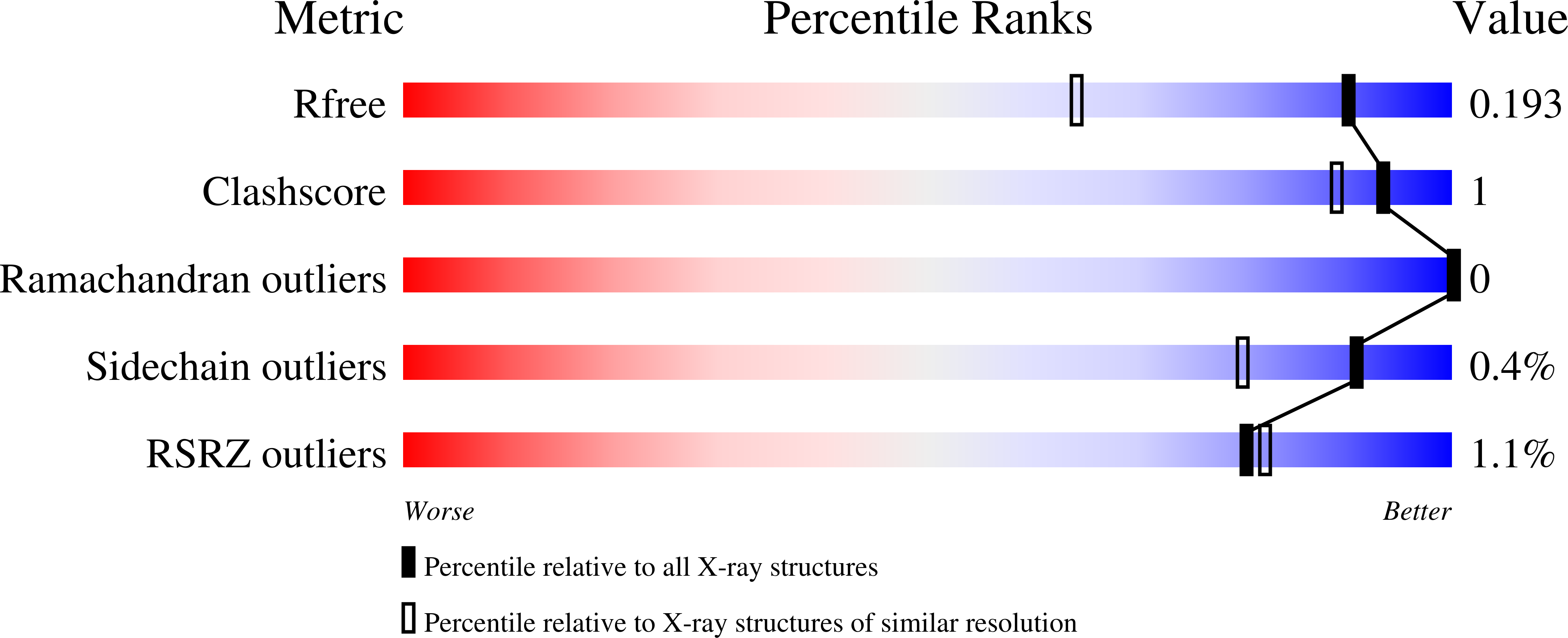
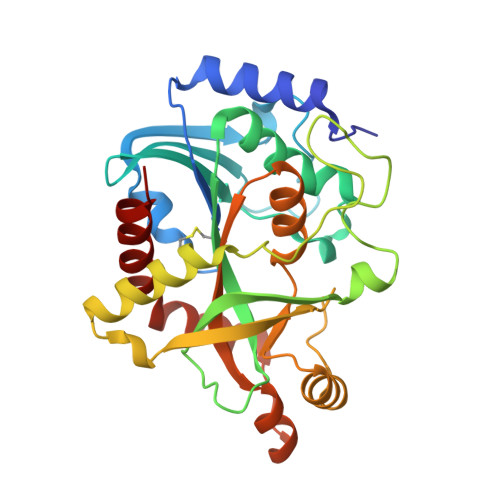
Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 3-methylpyridazin-1-ium-6-olate
Faheem, M., Neto, J.B., Collins, P., Pearce, N.M., Valadares, N.F., Bird, L., Pereira, H.M., Delft, F.V., Barbosa, J.A.R.G.To be published.