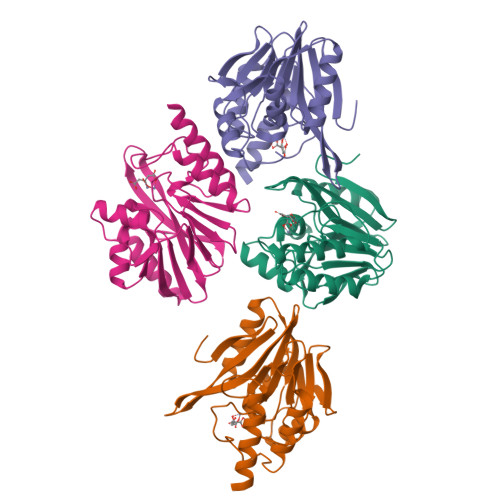
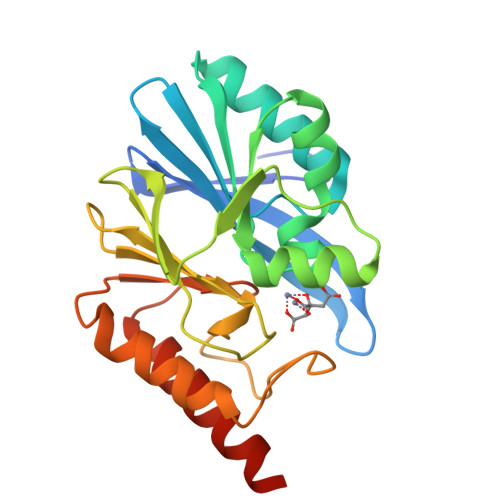
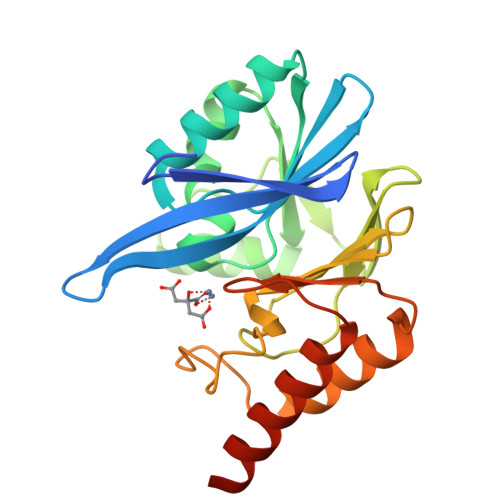
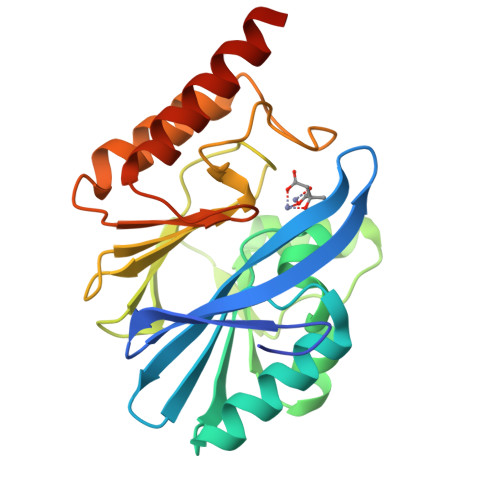
Genetic and Biochemical Characterization of VMB-1, a Novel Metallo-beta-Lactamase Encoded by a Conjugative, Broad-Host Range IncC Plasmid from Vibrio spp.
Zheng, Z., Cheng, Q., Chan, E.W., Chen, S.(2020) Adv Biosyst 4: e1900221-e1900221
- PubMed: 32293144
- DOI: https://doi.org/10.1002/adbi.201900221
- Primary Citation of Related Structures:
6JV4 - PubMed Abstract:
The increasing incidence of phenotypic resistance to carbapenems in recent years is mainly attributed to acquisition of mobile carbapenemase-encoding genetic elements by major bacterial pathogens. Here, a novel carbapenemase known as Vibrio metallo-β-lactamase 1 (VMB-1), which is encoded by a gene (bla VMB-1 ) located in an integron-bearing, highly transmissible IncC type plasmid, namely pVB1796, is identified and characterized, both genetically and functionally. Recovered from a foodborne Vibrio alginolyticus strain that exhibits resistance to all known β-lactam antibiotics, pVB1796 is found to possess a hybrid backbone that exhibits unique features of both type 1 and type 2 IncC elements. VMB-1 exhibits 94% sequence homology with several recently reported but poorly characterized metallo-β-lactamases (MBLs) produced by the marine organisms Alteromonadaceae, Glaciecola, and Thalassomonas actiniarum. Sequence alignment analysis shows that VMB-1 shares a structurally identical active site with subclass B1 MBLs. Importantly, pVB1796 is found to be efficiently transferred from Vibrio to other Gram-negative bacterial pathogens, including Salmonella typhimurium, Klebsiella pneumoniae, and Acinetobacter baumanni, via conjugation. These findings suggest that bla VMB-1 -bearing plasmids have the potential to be disseminated to other Gram-negative bacterial pathogens in the near future and render carbapenems useless in treatment of multidrug resistant infections.
Organizational Affiliation:
Shenzhen Key Laboratory for Food Biological Safety Control, Food Safety and Technology Research Centre, The Hong Kong PolyU Shenzhen Research Institute, Shenzhen, 518052, P. R. China.