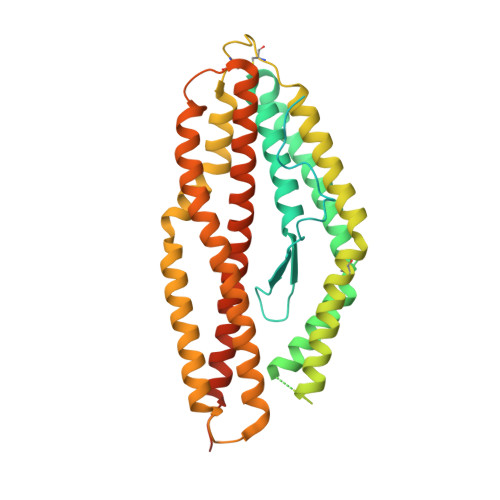
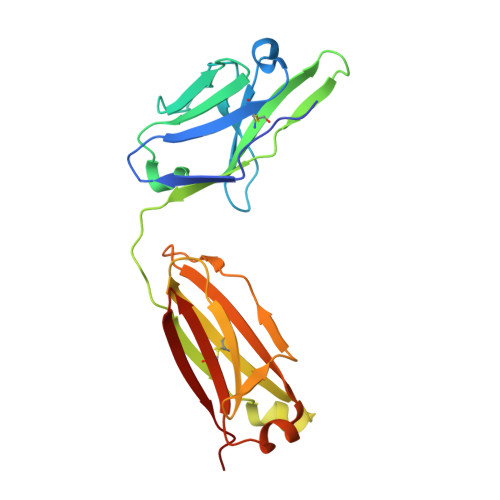
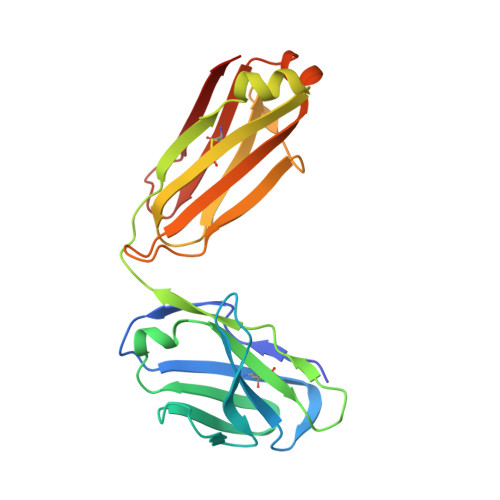
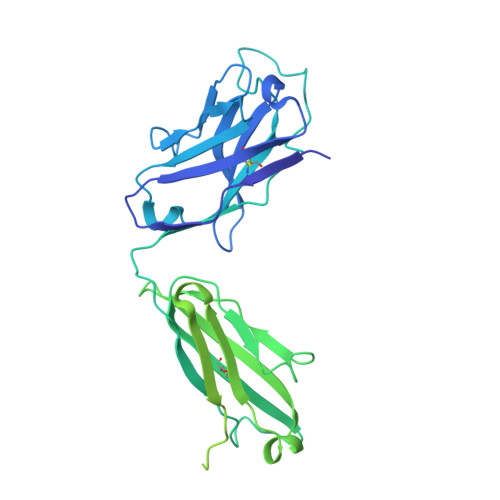
Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Alanine, D.G.W., Quinkert, D., Kumarasingha, R., Mehmood, S., Donnellan, F.R., Minkah, N.K., Dadonaite, B., Diouf, A., Galaway, F., Silk, S.E., Jamwal, A., Marshall, J.M., Miura, K., Foquet, L., Elias, S.C., Labbe, G.M., Douglas, A.D., Jin, J., Payne, R.O., Illingworth, J.J., Pattinson, D.J., Pulido, D., Williams, B.G., de Jongh, W.A., Wright, G.J., Kappe, S.H.I., Robinson, C.V., Long, C.A., Crabb, B.S., Gilson, P.R., Higgins, M.K., Draper, S.J.(2019) Cell 178: 216-228.e21
- PubMed: 31204103
- DOI: https://doi.org/10.1016/j.cell.2019.05.025
- Primary Citation of Related Structures:
6RCO, 6RCQ, 6RCS, 6RCU, 6RCV - PubMed Abstract:
The Plasmodium falciparum reticulocyte-binding protein homolog 5 (PfRH5) is the leading target for next-generation vaccines against the disease-causing blood-stage of malaria. However, little is known about how human antibodies confer functional immunity against this antigen. We isolated a panel of human monoclonal antibodies (mAbs) against PfRH5 from peripheral blood B cells from vaccinees in the first clinical trial of a PfRH5-based vaccine. We identified a subset of mAbs with neutralizing activity that bind to three distinct sites and another subset of mAbs that are non-functional, or even antagonistic to neutralizing antibodies. We also identify the epitope of a novel group of non-neutralizing antibodies that significantly reduce the speed of red blood cell invasion by the merozoite, thereby potentiating the effect of all neutralizing PfRH5 antibodies as well as synergizing with antibodies targeting other malaria invasion proteins. Our results provide a roadmap for structure-guided vaccine development to maximize antibody efficacy against blood-stage malaria.
Organizational Affiliation:
The Jenner Institute, University of Oxford, Old Road Campus Research Building, Oxford OX3 7DQ, UK; Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.