A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
Noske, G.D., Nakamura, A.M., Gawriljuk, V.O., Fernandes, R.S., Lima, G.M.A., Rosa, H.V.D., Pereira, H.D., Zeri, A.C.M., Nascimento, A.F.Z., Freire, M.C.L.C., Fearon, D., Douangamath, A., von Delft, F., Oliva, G., Godoy, A.S.(2021) J Mol Biol 433: 167118-167118
- PubMed: 34174328
- DOI: https://doi.org/10.1016/j.jmb.2021.167118
- Primary Citation of Related Structures:
7KFI, 7KPH, 7KVL, 7KVR, 7LDX, 7LFE, 7LFP, 7MBG, 7N5Z, 7N6N - PubMed Abstract:
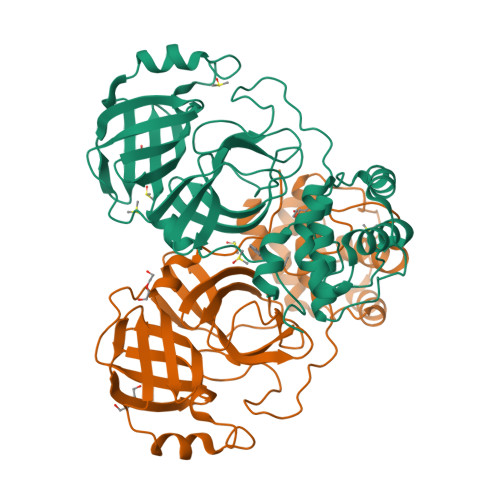
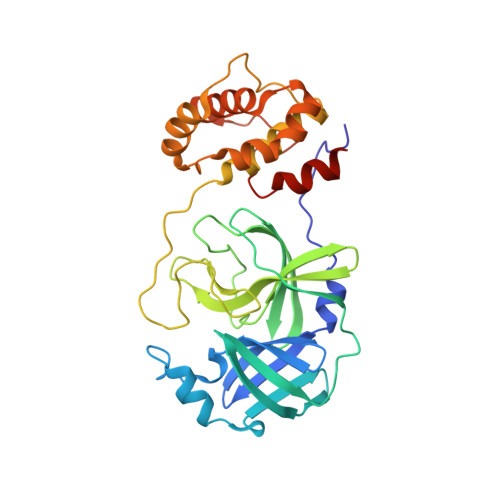
SARS-CoV-2 is the causative agent of COVID-19. The dimeric form of the viral M pro is responsible for the cleavage of the viral polyprotein in 11 sites, including its own N and C-terminus. The lack of structural information for intermediary forms of M pro is a setback for the understanding its self-maturation process. Herein, we used X-ray crystallography combined with biochemical data to characterize multiple forms of SARS-CoV-2 M pro . For the immature form, we show that extra N-terminal residues caused conformational changes in the positioning of domain-three over the active site, hampering the dimerization and diminishing its activity. We propose that this form preludes the cis and trans-cleavage of N-terminal residues. Using fragment screening, we probe new cavities in this form which can be used to guide therapeutic development. Furthermore, we characterized a serine site-directed mutant of the M pro bound to its endogenous N and C-terminal residues during dimeric association stage of the maturation process. We suggest this form is a transitional state during the C-terminal trans-cleavage. This data sheds light in the structural modifications of the SARS-CoV-2 main protease during its self-maturation process.
Organizational Affiliation:
Institute of Physics of Sao Carlos, University of Sao Paulo, Av. Joao Dagnone, 1100, Jardim Santa Angelina, Sao Carlos 13563-120, Brazil.