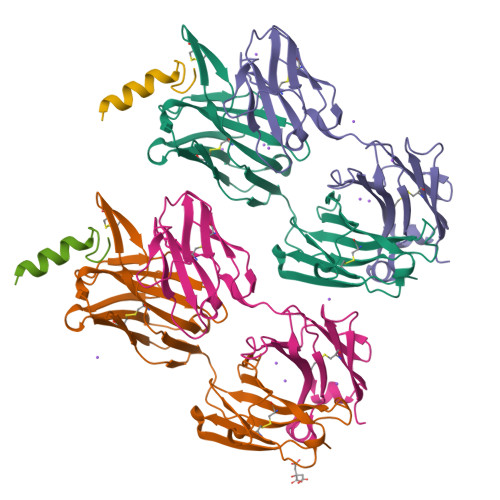
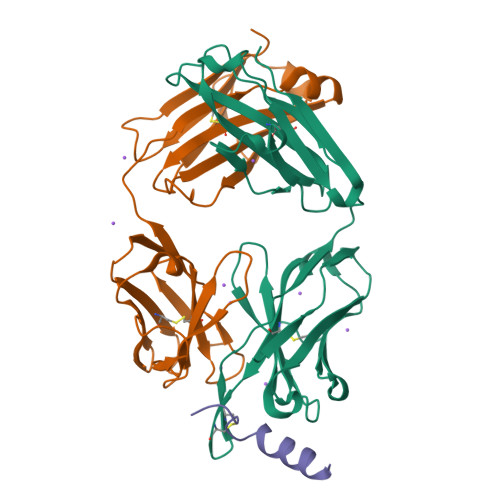
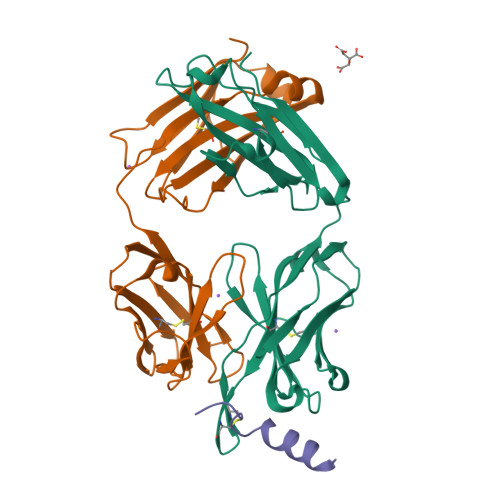
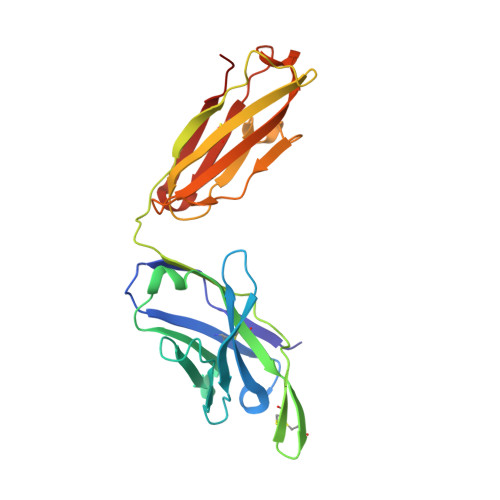
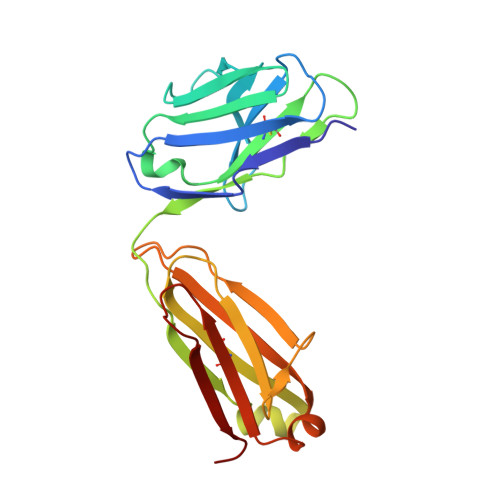
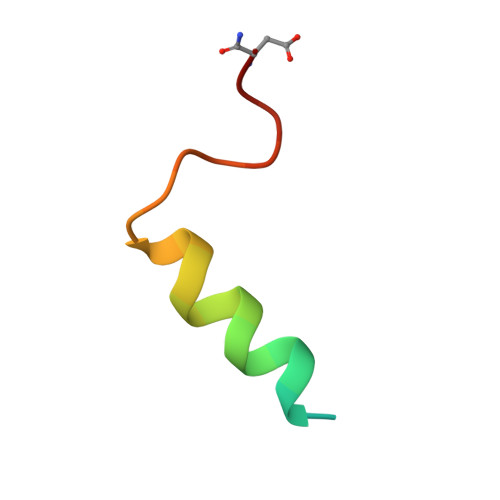
Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Li, W., Chen, Y., Prevost, J., Ullah, I., Lu, M., Gong, S.Y., Tauzin, A., Gasser, R., Vezina, D., Anand, S.P., Goyette, G., Chaterjee, D., Ding, S., Tolbert, W.D., Grunst, M.W., Bo, Y., Zhang, S., Richard, J., Zhou, F., Huang, R.K., Esser, L., Zeher, A., Cote, M., Kumar, P., Sodroski, J., Xia, D., Uchil, P.D., Pazgier, M., Finzi, A., Mothes, W.(2022) Cell Rep 38: 110210-110210
- PubMed: 34971573
- DOI: https://doi.org/10.1016/j.celrep.2021.110210
- Primary Citation of Related Structures:
7NAB - PubMed Abstract:
Emerging variants of concern for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can transmit more efficiently and partially evade protective immune responses, thus necessitating continued refinement of antibody therapies and immunogen design. Here, we elucidate the structural basis and mode of action for two potent SARS-CoV-2 spike (S)-neutralizing monoclonal antibodies, CV3-1 and CV3-25, which remain effective against emerging variants of concern in vitro and in vivo. CV3-1 binds to the (485-GFN-487) loop within the receptor-binding domain (RBD) in the "RBD-up" position and triggers potent shedding of the S1 subunit. In contrast, CV3-25 inhibits membrane fusion by binding to an epitope in the stem helix region of the S2 subunit that is highly conserved among β-coronaviruses. Thus, vaccine immunogen designs that incorporate the conserved regions in the RBD and stem helix region are candidates to elicit pan-coronavirus protective immune responses.
Organizational Affiliation:
Department of Microbial Pathogenesis, Yale University School of Medicine, New Haven, CT 06520, USA.