Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii
Rathbone, H.W., Curmi, P.M.G., Michie, K.A., Green, B.R., Laos, A.L., Thordarson, P., Iranmanesh, H.(2023) Commun Biol
Experimental Data Snapshot
(2023) Commun Biol
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HaPE560 alpha subunit | 72 | Hemiselmis andersenii | Mutation(s): 0 | 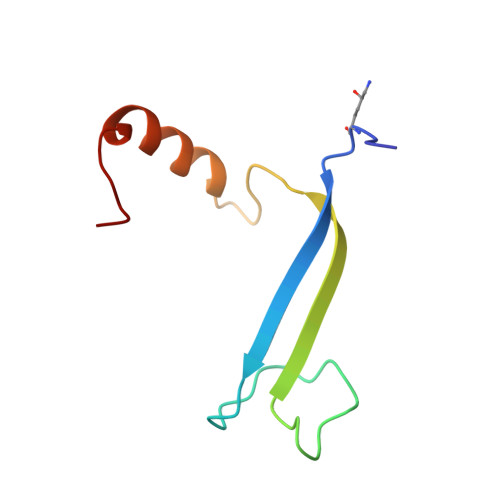 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phycoerythrin550 beta subunit | 177 | Hemiselmis andersenii | Mutation(s): 0 | 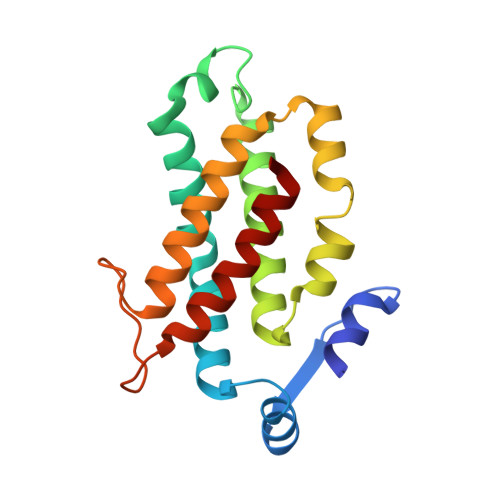 | |
UniProt | |||||
Find proteins for U5T8W0 (Hemiselmis andersenii) Explore U5T8W0 Go to UniProtKB: U5T8W0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | U5T8W0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AX9 (Subject of Investigation/LOI) Query on AX9 | J [auth B], O [auth D], S [auth F], Y [auth H] | DiCys-(15,16)-Dihydrobiliverdin C33 H40 N4 O6 MZFCOERRVCGRTL-ZTYGKHTCSA-N |  | ||
| PEB (Subject of Investigation/LOI) Query on PEB | AA [auth H] I [auth A] K [auth B] L [auth B] N [auth C] | PHYCOERYTHROBILIN C33 H40 N4 O6 NKCBCVIFPXGHAV-WAVSMFBNSA-N |  | ||
| TRS Query on TRS | BA [auth H], M [auth B], V [auth F] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| CL Query on CL | W [auth F] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| LYZ Query on LYZ | A, C, E, G | L-PEPTIDE LINKING | C6 H14 N2 O3 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 84.32 | α = 90 |
| b = 67.97 | β = 99.33 |
| c = 184.494 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| iMOSFLM | data reduction |
| pointless | data scaling |
| Aimless | data scaling |
| PHENIX | phasing |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Australian Research Council (ARC) | Australia | DP180103964 |
| Other government | Australia | FA2386-17-1-4101 (U.S. Air Force Office of Scientific Research through the Asian Office of Aerospace Research and Developmen) |
| Australian Research Council (ARC) | Australia | LE190100165 |