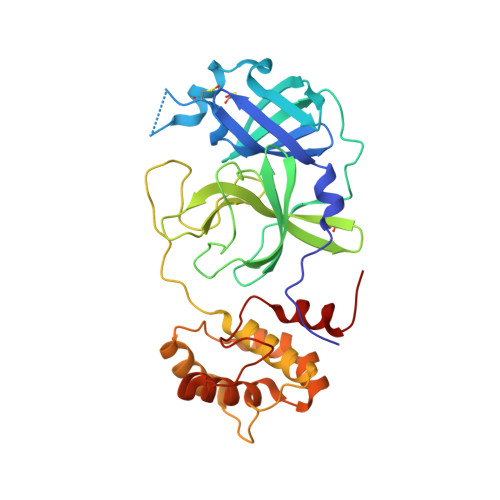
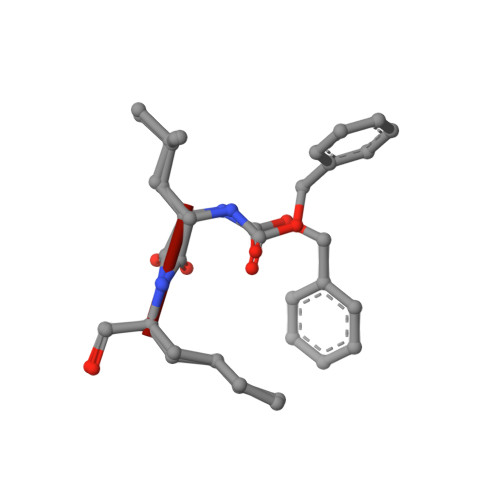
Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Reinke, P.Y.A., de Souza, E.E., Gunther, S., Falke, S., Lieske, J., Ewert, W., Loboda, J., Herrmann, A., Rahmani Mashhour, A., Karnicar, K., Usenik, A., Lindic, N., Sekirnik, A., Botosso, V.F., Santelli, G.M.M., Kapronezai, J., de Araujo, M.V., Silva-Pereira, T.T., Filho, A.F.S., Tavares, M.S., Florez-Alvarez, L., de Oliveira, D.B.L., Durigon, E.L., Giaretta, P.R., Heinemann, M.B., Hauser, M., Seychell, B., Bohler, H., Rut, W., Drag, M., Beck, T., Cox, R., Chapman, H.N., Betzel, C., Brehm, W., Hinrichs, W., Ebert, G., Latham, S.L., Guimaraes, A.M.S., Turk, D., Wrenger, C., Meents, A.(2023) Commun Biol 6: 1058-1058
- PubMed: 37853179
- DOI: https://doi.org/10.1038/s42003-023-05317-9
- Primary Citation of Related Structures:
7QGW, 7QKA, 7QKB, 7QKC, 7Z3T, 7Z3U, 7Z58, 8C3D - PubMed Abstract:
Several drug screening campaigns identified Calpeptin as a drug candidate against SARS-CoV-2. Initially reported to target the viral main protease (M pro ), its moderate activity in M pro inhibition assays hints at a second target. Indeed, we show that Calpeptin is an extremely potent cysteine cathepsin inhibitor, a finding additionally supported by X-ray crystallography. Cell infection assays proved Calpeptin's efficacy against SARS-CoV-2. Treatment of SARS-CoV-2-infected Golden Syrian hamsters with sulfonated Calpeptin at a dose of 1 mg/kg body weight reduces the viral load in the trachea. Despite a higher risk of side effects, an intrinsic advantage in targeting host proteins is their mutational stability in contrast to highly mutable viral targets. Here we show that the inhibition of cathepsins, a protein family of the host organism, by calpeptin is a promising approach for the treatment of SARS-CoV-2 and potentially other viral infections.
Organizational Affiliation:
Center for Free-Electron Laser Science CFEL, Deutsches Elektronen-Synchrotron DESY, Notkestr. 85, 22607, Hamburg, Germany.