Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Higashi-Kuwata, N., Tsuji, K., Hayashi, H., Bulut, H., Kiso, M., Imai, M., Ogata-Aoki, H., Ishii, T., Kobayakawa, T., Nakano, K., Takamune, N., Kishimoto, N., Hattori, S.I., Das, D., Uemura, Y., Shimizu, Y., Aoki, M., Hasegawa, K., Suzuki, S., Nishiyama, A., Saruwatari, J., Shimizu, Y., Sukenaga, Y., Takamatsu, Y., Tsuchiya, K., Maeda, K., Yoshimura, K., Iida, S., Ozono, S., Suzuki, T., Okamura, T., Misumi, S., Kawaoka, Y., Tamamura, H., Mitsuya, H.(2023) Nat Commun 14: 1076-1076
- PubMed: 36841831
- DOI: https://doi.org/10.1038/s41467-023-36729-0
- Primary Citation of Related Structures:
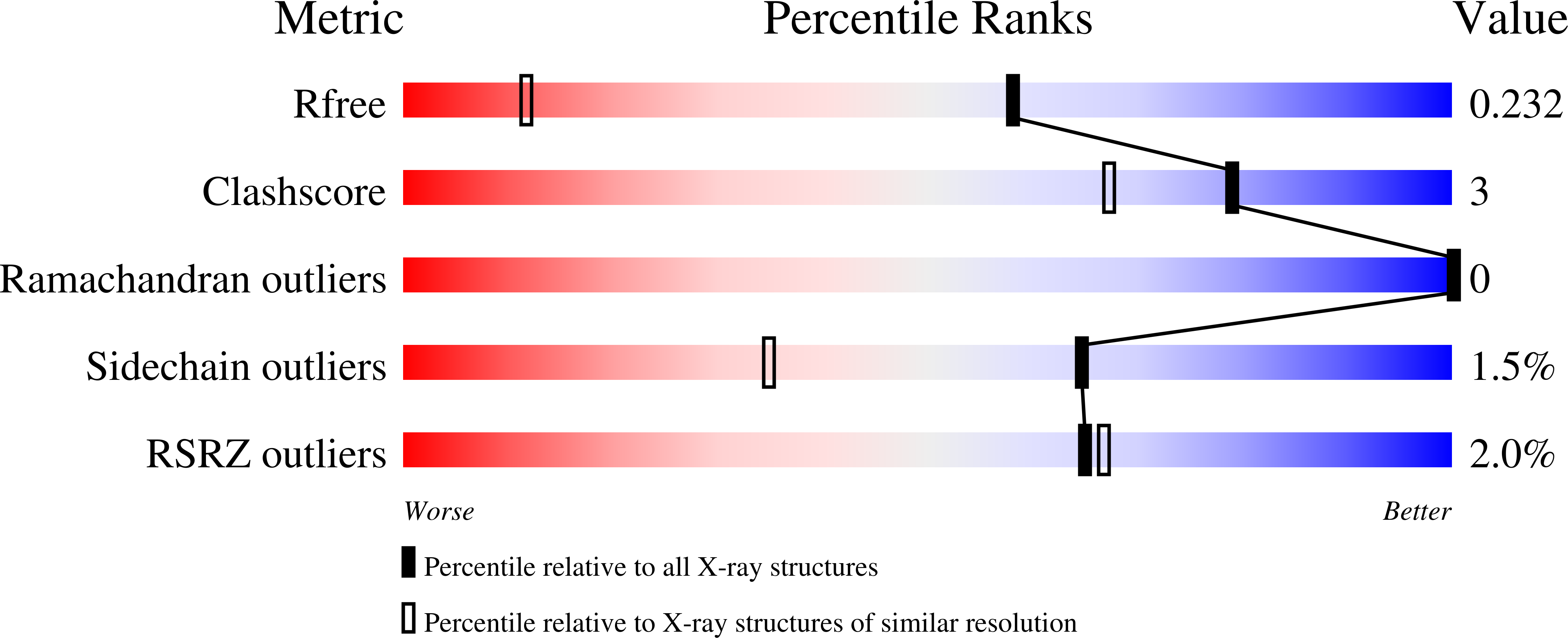
8DOX, 8DPR - PubMed Abstract:
COVID-19 caused by SARS-CoV-2 has continually been serious threat to public health worldwide. While a few anti-SARS-CoV-2 therapeutics are currently available, their antiviral potency is not sufficient. Here, we identify two orally available 4-fluoro-benzothiazole-containing small molecules, TKB245 and TKB248, which specifically inhibit the enzymatic activity of main protease (M pro ) of SARS-CoV-2 and significantly more potently block the infectivity and replication of various SARS-CoV-2 strains than nirmatrelvir, molnupiravir, and ensitrelvir in cell-based assays employing various target cells. Both compounds also block the replication of Delta and Omicron variants in human-ACE2-knocked-in mice. Native mass spectrometric analysis reveals that both compounds bind to dimer M pro , apparently promoting M pro dimerization. X-ray crystallographic analysis shows that both compounds bind to M pro 's active-site cavity, forming a covalent bond with the catalytic amino acid Cys-145 with the 4-fluorine of the benzothiazole moiety pointed to solvent. The data suggest that TKB245 and TKB248 might serve as potential therapeutics for COVID-19 and shed light upon further optimization to develop more potent and safer anti-SARS-CoV-2 therapeutics.
Organizational Affiliation:
Department of Refractory Viral Diseases, National Center for Global Health and Medicine Research Institute, Tokyo, Japan.