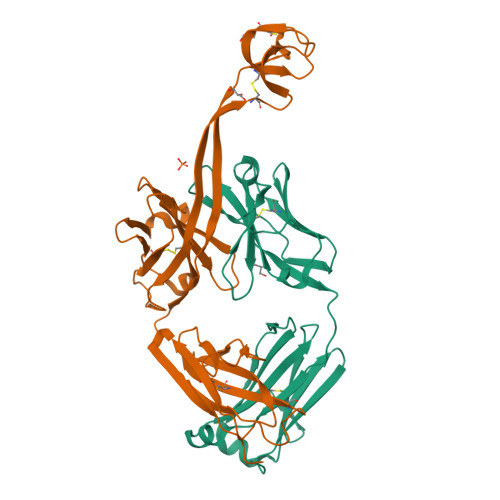
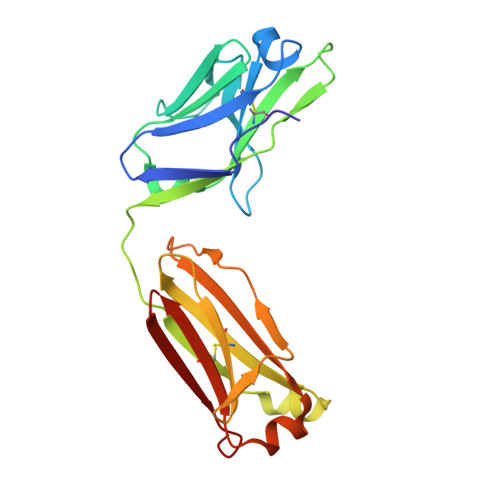
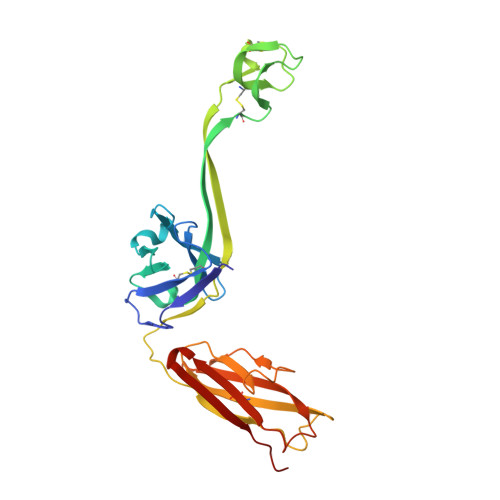
The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Huang, R., Warner Jenkins, G., Kim, Y., Stanfield, R.L., Singh, A., Martinez-Yamout, M., Kroon, G.J., Torres, J.L., Jackson, A.M., Kelley, A., Shaabani, N., Zeng, B., Bacica, M., Chen, W., Warner, C., Radoicic, J., Joh, J., Dinali Perera, K., Sang, H., Kim, T., Yao, J., Zhao, F., Sok, D., Burton, D.R., Allen, J., Harriman, W., Mwangi, W., Chung, D., Teijaro, J.R., Ward, A.B., Dyson, H.J., Wright, P.E., Wilson, I.A., Chang, K.O., McGregor, D., Smider, V.V.(2023) Proc Natl Acad Sci U S A 120: e2303455120-e2303455120
- PubMed: 37722054
- DOI: https://doi.org/10.1073/pnas.2303455120
- Primary Citation of Related Structures:
8ECQ, 8ECV, 8ECZ, 8ED1, 8EDF - PubMed Abstract:
Cows produce antibodies with a disulfide-bonded antigen-binding domain embedded within ultralong heavy chain third complementarity determining regions. This "knob" domain is analogous to natural cysteine-rich peptides such as knottins in that it is small and stable but can accommodate diverse loops and disulfide bonding patterns. We immunized cattle with SARS-CoV-2 spike and found ultralong CDR H3 antibodies that could neutralize several viral variants at picomolar IC 50 potencies in vitro and could protect from disease in vivo. The independent CDR H3 peptide knobs were expressed and maintained the properties of the parent antibodies. The knob interaction with SARS-CoV-2 spike was revealed by electron microscopy, X-ray crystallography, NMR spectroscopy, and mass spectrometry and established ultralong CDR H3-derived knobs as the smallest known recombinant independent antigen-binding fragment. Unlike other vertebrate antibody fragments, these knobs are not reliant on the immunoglobulin domain and have potential as a new class of therapeutics.
Organizational Affiliation:
Applied Biomedical Science Institute, San Diego, CA 92127.