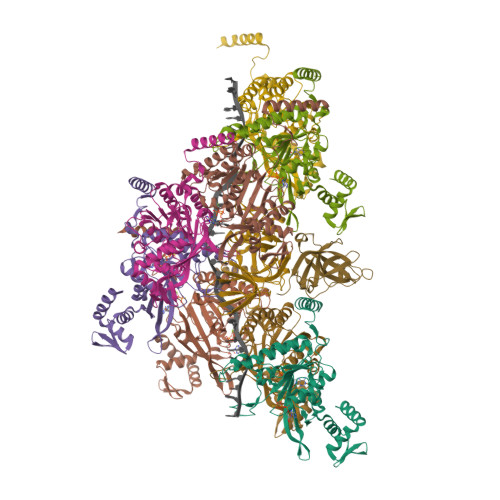
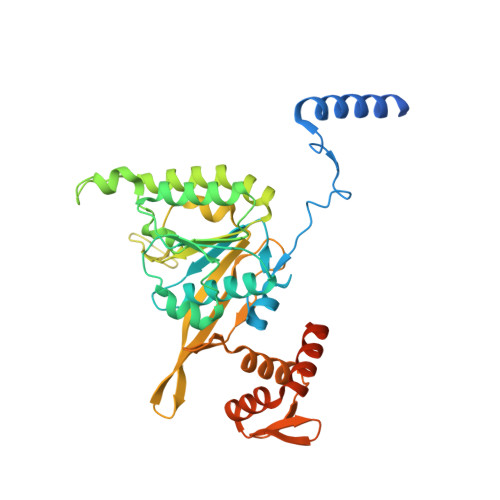
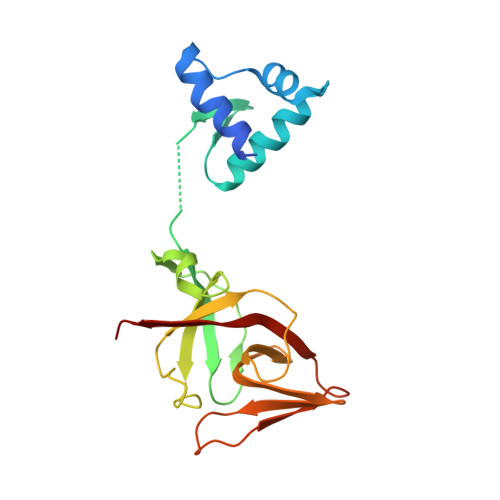
The LexA-RecA* structure reveals a cryptic lock-and-key mechanism for SOS activation.
Cory, M.B., Li, A., Hurley, C.M., Carman, P.J., Pumroy, R.A., Hostetler, Z.M., Perez, R.M., Venkatesh, Y., Li, X., Gupta, K., Petersson, E.J., Kohli, R.M.(2024) Nat Struct Mol Biol 31: 1522-1531
- PubMed: 38755298
- DOI: https://doi.org/10.1038/s41594-024-01317-3
- Primary Citation of Related Structures:
8TRG - PubMed Abstract:
The bacterial SOS response plays a key role in adaptation to DNA damage, including genomic stress caused by antibiotics. SOS induction begins when activated RecA*, an oligomeric nucleoprotein filament that forms on single-stranded DNA, binds to and stimulates autoproteolysis of the repressor LexA. Here, we present the structure of the complete Escherichia coli SOS signal complex, constituting full-length LexA bound to RecA*. We uncover an extensive interface unexpectedly including the LexA DNA-binding domain, providing a new molecular rationale for ordered SOS gene induction. We further find that the interface involves three RecA subunits, with a single residue in the central engaged subunit acting as a molecular key, inserting into an allosteric binding pocket to induce LexA cleavage. Given the pro-mutagenic nature of SOS activation, our structural and mechanistic insights provide a foundation for developing new therapeutics to slow the evolution of antibiotic resistance.
Organizational Affiliation:
Graduate Group in Biochemistry and Biophysics, University of Pennsylvania, Philadelphia, PA, USA.