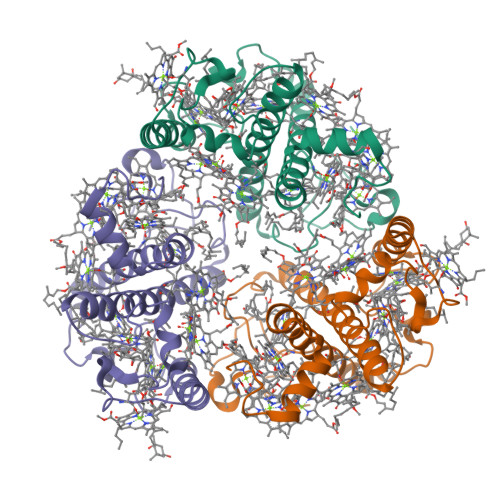
Structure-based validation of recombinant light-harvesting complex II.
Seki, S., Miyata, T., Norioka, N., Tanaka, H., Kurisu, G., Namba, K., Fujii, R.(2024) PNAS Nexus 3: pgae405-pgae405
- PubMed: 39346626
- DOI: https://doi.org/10.1093/pnasnexus/pgae405
- Primary Citation of Related Structures:
8Y15, 8YEE - PubMed Abstract:
Light-harvesting complex II (LHCII) captures sunlight and dissipates excess energy to drive photosynthesis. To elucidate this mechanism, the individual optical properties of pigments in the LHCII protein must be identified. In vitro reconstitution with apoproteins synthesized by Escherichia coli and pigment-lipid mixtures from natural sources is an effective approach; however, the local environment surrounding each pigment within reconstituted LHCII (rLHCII) has only been indirectly estimated using spectroscopic and biochemical methods. Here, we used cryo-electron microscopy to determine the 3D structure of the rLHCII trimer and found that rLHCII exhibited a structure that was virtually identical to that of native LHCII, with a few exceptions: some C-terminal amino acids were not visible, likely due to aggregation of the His-tags; a carotenoid at the V1 site was not visible; and at site 614 showed mixed occupancy by both chlorophyll a and b molecules. Our observations confirmed the applicability of the in vitro reconstitution technique.
Organizational Affiliation:
Graduate School of Science, Osaka City University, 3-3-138 Sugimoto, Sumiyoshi-ku, Osaka 558-8585, Japan.