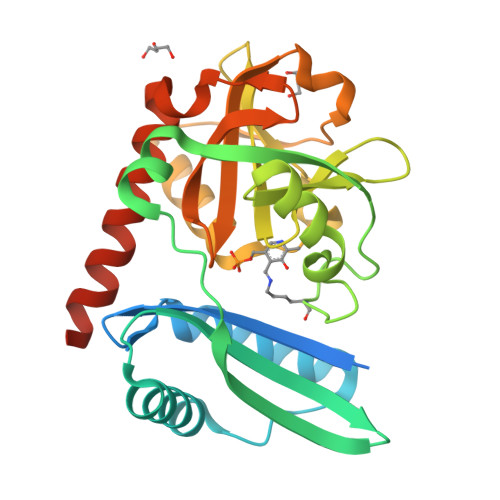
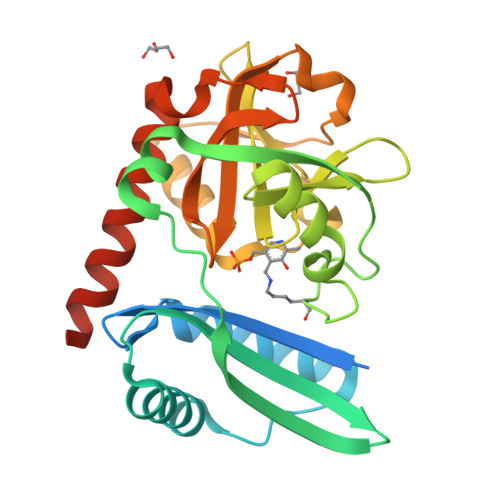
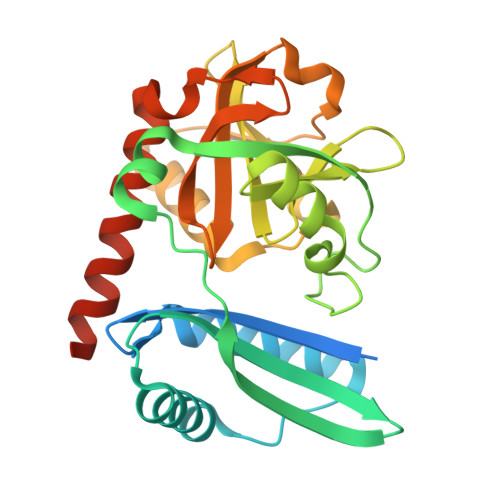
From Structure to Function: Analysis of the First Monomeric Pyridoxal-5'-Phosphate-Dependent Transaminase from the Bacterium Desulfobacula toluolica .
Bakunova, A.K., Matyuta, I.O., Nikolaeva, A.Y., Rakitina, T.V., Boyko, K.M., Popov, V.O., Bezsudnova, E.Y.(2024) Biomolecules 14
- PubMed: 39766298
- DOI: https://doi.org/10.3390/biom14121591
- Primary Citation of Related Structures:
9J0U, 9J0V - PubMed Abstract:
The first monomeric pyridoxal-5'-phosphate (PLP)-dependent transaminase from a marine, aromatic-compound-degrading, sulfate-reducing bacterium Desulfobacula toluolica Tol2, has been studied using structural, kinetic, and spectral methods. The monomeric organization of the transaminase was confirmed by both gel filtration and crystallography. The PLP-dependent transaminase is of the fold type IV and deaminates D-alanine and ( R )-phenylethylamine in half-reactions. The enzyme shows high stereoselectivity; no deamination of L-amino acids and ( S )-phenylethylamine is detected. Structural analysis and subsequent mutagenesis led to the conclusion that the monomeric architecture of the enzyme is the only one possible and sufficient for stereoselectivity and PLP binding, but not for the overall double-substrate transamination reaction and the stability of the holo form with the reduced cofactor-pyridoxamine-5'-phosphate. These results extend the structural university of the PLP fold type IV enzymes and demonstrate the need for deeper analysis of the sequence-structure-function relationships in the transaminases.
Organizational Affiliation:
Bach Institute of Biochemistry, Research Centre of Biotechnology of the Russian Academy of Sciences, Leninsky Ave. 33, bld. 2, 119071 Moscow, Russia.