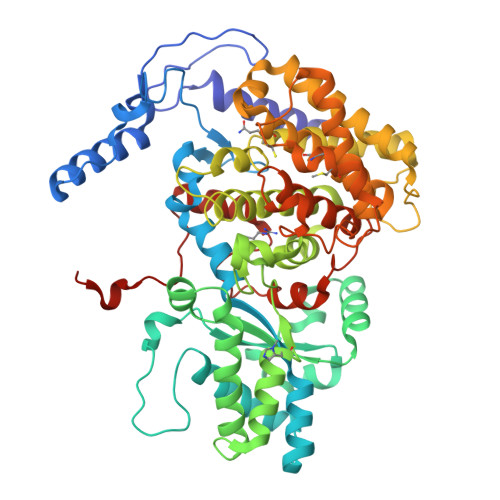
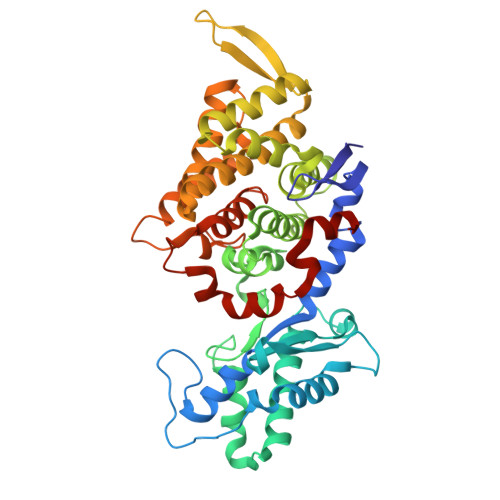
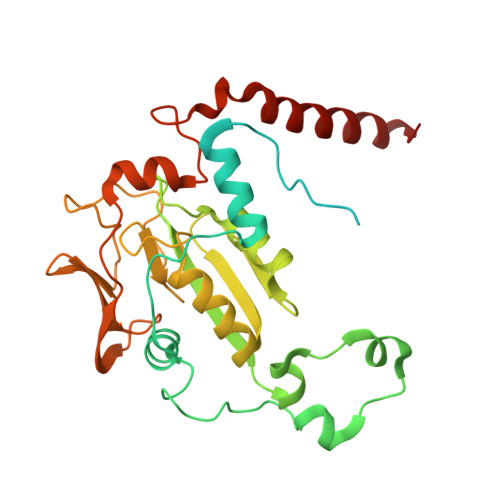
Genetic and biochemical characterization of a radical SAM enzyme required for posttranslational glutamine methylation of methyl-coenzyme M reductase
Rodriguez Carrero, R.J., Lloyd, C.T., Borkar, J., Nath, S., Mirica, L.M., Nair, S.K., Booker, S.J., Metcalf, W.To be published.